Distinct effects of concomitant Jak2V617F expression and Tet2 loss in mice promote disease progression in myeloproliferative neoplasms
- PMID: 25281607
- PMCID: PMC4287639
- DOI: 10.1182/blood-2014-04-567024
Distinct effects of concomitant Jak2V617F expression and Tet2 loss in mice promote disease progression in myeloproliferative neoplasms
Abstract
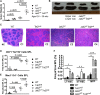
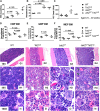
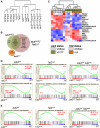
Signaling mutations (eg, JAK2V617F) and mutations in genes involved in epigenetic regulation (eg, TET2) are the most common cooccurring classes of mutations in myeloproliferative neoplasms (MPNs). Clinical correlative studies have demonstrated that TET2 mutations are enriched in more advanced phases of MPNs such as myelofibrosis and leukemic transformation, suggesting that they may cooperate with JAK2V617F to promote disease progression. To dissect the effects of concomitant Jak2V617F expression and Tet2 loss within distinct hematopoietic compartments in vivo, we generated Jak2V617F/Tet2 compound mutant genetic mice. We found that the combination of Jak2V617F expression and Tet2 loss resulted in a more florid MPN phenotype than that seen with either allele alone. Concordant with this, we found that Tet2 deletion conferred a strong functional competitive advantage to Jak2V617F-mutant hematopoietic stem cells (HSCs). Transcriptional profiling revealed that both Jak2V617F expression and Tet2 loss were associated with distinct and nonoverlapping gene expression signatures within the HSC compartment. In aggregate, our findings indicate that Tet2 loss drives clonal dominance in HSCs, and Jak2V617F expression causes expansion of downstream precursor cell populations, resulting in disease progression through combinatorial effects. This work provides insight into the functional consequences of JAK2V617F-TET2 comutation in MPNs, particularly as it pertains to HSCs.
© 2015 by The American Society of Hematology.
Figures

Comment in
-
TET2 loss, a rescue of JAK2V617F HSCs.Blood. 2015 Jan 8;125(2):212-3. doi: 10.1182/blood-2014-10-606624. Blood. 2015. PMID: 25573967
References
-
- Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA. New mutations and pathogenesis of myeloproliferative neoplasms. Blood. 2011;118(7):1723–1735. - PubMed
-
- Baxter EJ, Scott LM, Campbell PJ, et al. Cancer Genome Project. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365(9464):1054–1061. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

