CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis
- PMID: 25281690
- PMCID: PMC4247566
- DOI: 10.1105/tpc.114.128611
CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis
Abstract
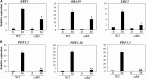
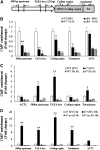
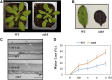
CYCLIN-DEPENDENT KINASE8 (CDK8) is a widely studied component of eukaryotic Mediator complexes. However, the biological and molecular functions of plant CDK8 are not well understood. Here, we provide evidence for regulatory functions of Arabidopsis thaliana CDK8 in defense and demonstrate its functional and molecular interactions with other Mediator and non-Mediator subunits. The cdk8 mutant exhibits enhanced resistance to Botrytis cinerea but susceptibility to Alternaria brassicicola. The contributions of CDK8 to the transcriptional activation of defensin gene PDF1.2 and its interaction with MEDIATOR COMPLEX SUBUNIT25 (MED25) implicate CDK8 in jasmonate-mediated defense. Moreover, CDK8 associates with the promoter of AGMATINE COUMAROYLTRANSFERASE to promote its transcription and regulate the biosynthesis of the defense-active secondary metabolites hydroxycinnamic acid amides. CDK8 also interacts with the transcription factor WAX INDUCER1, implying its additional role in cuticle development. In addition, overlapping functions of CDK8 with MED12 and MED13 and interactions between CDK8 and C-type cyclins suggest the conserved configuration of the plant Mediator kinase module. In summary, while CDK8's positive transcriptional regulation of target genes and its phosphorylation activities underpin its defense functions, the impaired defense responses in the mutant are masked by its altered cuticle, resulting in specific resistance to B. cinerea.
© 2014 American Society of Plant Biologists. All rights reserved.
Figures








References
-
- AbuQamar S., Chen X., Dhawan R., Bluhm B., Salmeron J., Lam S., Dietrich R.A., Mengiste T. (2006). Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 48: 28–44. - PubMed
-
- Andrau J.C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M.G., van de Peppel J., Werner M., Holstege F.C. (2006). Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22: 179–192. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

