Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor
- PMID: 25282148
- PMCID: PMC4222816
- DOI: 10.1038/nsmb.2899
Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor
Abstract
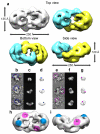
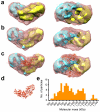
Mutations in BRCA2 increase susceptibility to breast, ovarian and prostate cancers. The product of human BRCA2, BRCA2 protein, has a key role in the repair of DNA double-strand breaks and interstrand cross-links by RAD51-mediated homologous recombination. Here, we present a biochemical and structural characterization of full-length (3,418 amino acid) BRCA2, alone and in complex with RAD51. We show that BRCA2 facilitates nucleation of RAD51 filaments at multiple sites on single-stranded DNA. Three-dimensional EM reconstructions revealed that BRCA2 exists as a dimer and that two oppositely oriented sets of RAD51 molecules bind the dimer. Single-stranded DNA binds along the long axis of BRCA2, such that only one set of RAD51 monomers can form a productive complex with DNA and establish filament formation. Our data define the molecular mechanism by which this tumor suppressor facilitates RAD51-mediated homologous-recombinational repair.
Figures



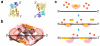
Similar articles
-
BRCA2 C-terminal clamp restructures RAD51 dimers to bind B-DNA for replication fork stability.Mol Cell. 2025 Jun 5;85(11):2080-2096.e6. doi: 10.1016/j.molcel.2025.05.010. Epub 2025 May 28. Mol Cell. 2025. PMID: 40441151
-
The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction.Nature. 2005 Feb 10;433(7026):653-7. doi: 10.1038/nature03234. Nature. 2005. PMID: 15703751
-
Dissecting the Recombination Mediator Activity of BRCA2 Using Biochemical Methods.Methods Enzymol. 2018;600:479-511. doi: 10.1016/bs.mie.2017.11.018. Methods Enzymol. 2018. PMID: 29458771
-
Unraveling the mechanism of BRCA2 in homologous recombination.Nat Struct Mol Biol. 2011 Jul 6;18(7):748-54. doi: 10.1038/nsmb.2096. Nat Struct Mol Biol. 2011. PMID: 21731065 Free PMC article. Review.
-
RecA: Regulation and Mechanism of a Molecular Search Engine.Trends Biochem Sci. 2016 Jun;41(6):491-507. doi: 10.1016/j.tibs.2016.04.002. Epub 2016 May 4. Trends Biochem Sci. 2016. PMID: 27156117 Free PMC article. Review.
Cited by
-
BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells.J Cell Biol. 2014 Dec 8;207(5):599-613. doi: 10.1083/jcb.201405014. J Cell Biol. 2014. PMID: 25488918 Free PMC article.
-
DSS1 and ssDNA regulate oligomerization of BRCA2.Nucleic Acids Res. 2020 Aug 20;48(14):7818-7833. doi: 10.1093/nar/gkaa555. Nucleic Acids Res. 2020. PMID: 32609828 Free PMC article.
-
Functional pre-therapeutic evaluation by genome editing of variants of uncertain significance of essential tumor suppressor genes.Genome Med. 2021 Nov 9;13(1):174. doi: 10.1186/s13073-021-00976-x. Genome Med. 2021. PMID: 34749799 Free PMC article.
-
Distinct implications of different BRCA mutations: efficacy of cytotoxic chemotherapy, PARP inhibition and clinical outcome in ovarian cancer.Onco Targets Ther. 2017 May 11;10:2539-2551. doi: 10.2147/OTT.S102569. eCollection 2017. Onco Targets Ther. 2017. PMID: 28546758 Free PMC article. Review.
-
Loss of the N-terminal methyltransferase NRMT1 increases sensitivity to DNA damage and promotes mammary oncogenesis.Oncotarget. 2015 May 20;6(14):12248-63. doi: 10.18632/oncotarget.3653. Oncotarget. 2015. PMID: 25909287 Free PMC article.
References
-
- Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 2008;40:17–22. - PubMed
-
- Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. - PubMed
-
- King MC, Marks JH, Mandell JB, Grp NYBCS. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

