A genome-wide map of adeno-associated virus-mediated human gene targeting
- PMID: 25282150
- PMCID: PMC4405182
- DOI: 10.1038/nsmb.2895
A genome-wide map of adeno-associated virus-mediated human gene targeting
Abstract
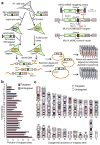
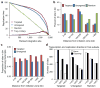
To determine which genomic features promote homologous recombination, we created a genome-wide map of gene targeting sites. We used an adeno-associated virus vector to target identical loci introduced as transcriptionally active retroviral vectors. A comparison of ~2,000 targeted and untargeted sites showed that targeting occurred throughout the human genome and was not influenced by the presence of nearby CpG islands, sequence repeats or DNase I-hypersensitive sites. Targeted sites were preferentially located within transcription units, especially when the target loci were transcribed in the opposite orientation to their surrounding chromosomal genes. We determined the impact of DNA replication by mapping replication forks, which revealed a preference for recombination at target loci transcribed toward an incoming fork. Our results constitute the first genome-wide screen of gene targeting in mammalian cells and demonstrate a strong recombinogenic effect of colliding polymerases.
Figures
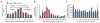

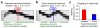

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

