Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials
- PMID: 25282355
- PMCID: PMC4317352
- DOI: 10.1038/nbt.3043
Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials
Abstract
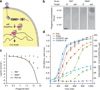
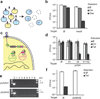
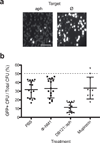
Antibiotics target conserved bacterial cellular pathways or growth functions and therefore cannot selectively kill specific members of a complex microbial population. Here, we develop programmable, sequence-specific antimicrobials using the RNA-guided nuclease Cas9 (refs.1,2) delivered by a bacteriophage. We show that Cas9, reprogrammed to target virulence genes, kills virulent, but not avirulent, Staphylococcus aureus. Reprogramming the nuclease to target antibiotic resistance genes destroys staphylococcal plasmids that harbor antibiotic resistance genes and immunizes avirulent staphylococci to prevent the spread of plasmid-borne resistance genes. We also show that CRISPR-Cas9 antimicrobials function in vivo to kill S. aureus in a mouse skin colonization model. This technology creates opportunities to manipulate complex bacterial populations in a sequence-specific manner.
Figures
Comment in
-
A CRISPR design for next-generation antimicrobials.Genome Biol. 2014 Nov 8;15(11):516. doi: 10.1186/s13059-014-0516-x. Genome Biol. 2014. PMID: 25417800 Free PMC article.
References
-
- Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. - PubMed
-
- Weigel LM, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

