Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells
- PMID: 25283993
- PMCID: PMC4298702
- DOI: 10.1038/ncb3045
Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells
Abstract
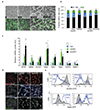
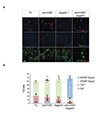
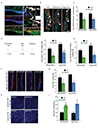
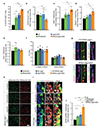
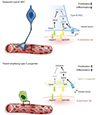
The vasculature is a prominent component of the subventricular zone neural stem cell niche. Although quiescent neural stem cells physically contact blood vessels at specialized endfeet, the significance of this interaction is not understood. In contrast, it is well established that vasculature-secreted soluble factors promote lineage progression of committed progenitors. Here we specifically investigated the role of cell-cell contact-dependent signalling in the vascular niche. Unexpectedly, we find that direct cell-cell interactions with endothelial cells enforce quiescence and promote stem cell identity. Mechanistically, endothelial ephrinB2 and Jagged1 mediate these effects by suppressing cell-cycle entry downstream of mitogens and inducing stemness genes to jointly inhibit differentiation. In vivo, endothelial-specific ablation of either of the genes which encode these proteins, Efnb2 and Jag1 respectively, aberrantly activates quiescent stem cells, resulting in depletion. Thus, we identify the vasculature as a critical niche compartment for stem cell maintenance, furthering our understanding of how anchorage to the niche maintains stem cells within a pro-differentiative microenvironment.
Figures



References
-
- Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–778. - PubMed
-
- Chen S, Wang S, Xie T. Restricting self-renewal signals within the stem cell niche: multiple levels of control. Current opinion in genetics & development. 2011;21:684–689. doi:10.1016/j.gde.2011.07.008. - PubMed
-
- Wagers AJ. The stem cell niche in regenerative medicine. Cell Stem Cell. 2012;10:362–369. doi:10.1016/j.stem.2012.02.018. - PubMed
-
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

