Multiple Mechanisms Contribute To Telomere Maintenance
- PMID: 25285314
- PMCID: PMC4181876
Multiple Mechanisms Contribute To Telomere Maintenance
Abstract
The unlimited growth potential of tumors depends on telomere maintenance and typically depends on telomerase, an RNA-dependent DNA polymerase, which reverse transcribes the telomerase RNA template, synthesizing telomere repeats at the ends of chromosomes. Studies in various model organisms genetically deleted for telomerase indicate that several recombination-based mechanisms also contribute to telomere maintenance. Understanding the molecular basis of these mechanisms is critical since some human tumors form without telomerase, yet the sequence is maintained at the telomeres. Recombination-based mechanisms also likely contribute at some frequency to telomere maintenance in tumors expressing telomerase. Preventing telomere maintenance is predicted to impact tumor growth, yet inhibiting telomerase may select for the recombination-based mechanisms. Telomere recombination mechanisms likely involve altered or unregulated pathways of DNA repair. The use of some DNA damaging agents may encourage the use of these unregulated pathways of DNA repair to be utilized and may allow some tumors to generate resistance to these agents depending on which repair pathways are altered in the tumors. This review will discuss the various telomere recombination mechanisms and will provide rationale regarding the possibility that L1 retrotransposition may contribute to telomere maintenance in tumors lacking telomerase.
Keywords: DNA repair; Retrotransposons; Telomere; Tumors.
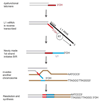
Figures



References
-
- Muller HJ. The remaking of chromosomes. Collecting Net. 1938;13:181.
-
- Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. - PubMed
-
- Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. - PubMed
-
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. - PubMed
-
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
