Next-generation sequencing analysis of miRNA expression in control and FSHD myogenesis
- PMID: 25285664
- PMCID: PMC4186784
- DOI: 10.1371/journal.pone.0108411
Next-generation sequencing analysis of miRNA expression in control and FSHD myogenesis
Abstract
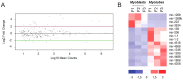
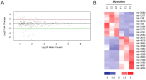
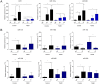
Emerging evidence has demonstrated that miRNA sequences can regulate skeletal myogenesis by controlling the process of myoblast proliferation and differentiation. However, at present a deep analysis of miRNA expression in control and FSHD myoblasts during differentiation has not yet been derived. To close this gap, we used a next-generation sequencing (NGS) approach applied to in vitro myogenesis. Furthermore, to minimize sample genetic heterogeneity and muscle-type specific patterns of gene expression, miRNA profiling from NGS data was filtered with FC ≥ 4 (log(2)FC ≥ 2) and p-value<0.05, and its validation was derived by qRT-PCR on myoblasts from seven muscle districts. In particular, control myogenesis showed the modulation of 38 miRNAs, the majority of which (34 out 38) were up-regulated, including myomiRs (miR-1, -133a, -133b and -206). Approximately one third of the modulated miRNAs were not previously reported to be involved in muscle differentiation, and interestingly some of these (i.e. miR-874, -1290, -95 and -146a) were previously shown to regulate cell proliferation and differentiation. FSHD myogenesis evidenced a reduced number of modulated miRNAs than healthy muscle cells. The two processes shared nine miRNAs, including myomiRs, although with FC values lower in FSHD than in control cells. In addition, FSHD cells showed the modulation of six miRNAs (miR-1268, -1268b, -1908, 4258, -4508- and -4516) not evidenced in control cells and that therefore could be considered FSHD-specific, likewise three novel miRNAs that seem to be specifically expressed in FSHD myotubes. These data further clarify the impact of miRNA regulation during control myogenesis and strongly suggest that a complex dysregulation of miRNA expression characterizes FSHD, impairing two important features of myogenesis: cell cycle and muscle development. The derived miRNA profiling could represent a novel molecular signature for FSHD that includes diagnostic biomarkers and possibly therapeutic targets.
Conflict of interest statement
Figures



References
-
- Padberg GW, Lunt PW, Koch M, Fardeau M (1992) Diagnostic criteria for facioscapulohumeral muscular dystrophy. Neuromuscul Disord 1: 231–4. - PubMed
-
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, et al. (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nature Genetics 2: 26–30. - PubMed
-
- Yamanaka G, Goto K, Ishihara T, Oya Y, Miyajima T, et al. (2004) FSHD-like patients without 4q35 deletion. J Neurol Sci 219: 89–93. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

