Double-strand break repair-adox: Restoration of suppressed double-strand break repair during mitosis induces genomic instability
- PMID: 25287622
- PMCID: PMC4317954
- DOI: 10.1111/cas.12551
Double-strand break repair-adox: Restoration of suppressed double-strand break repair during mitosis induces genomic instability
Abstract
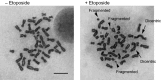
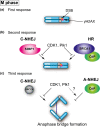
Double-strand breaks (DSBs) are one of the severest types of DNA damage. Unrepaired DSBs easily induce cell death and chromosome aberrations. To maintain genomic stability, cells have checkpoint and DSB repair systems to respond to DNA damage throughout most of the cell cycle. The failure of this process often results in apoptosis or genomic instability, such as aneuploidy, deletion, or translocation. Therefore, DSB repair is essential for maintenance of genomic stability. During mitosis, however, cells seem to suppress the DNA damage response and proceed to the next G1 phase, even if there are unrepaired DSBs. The biological significance of this suppression is not known. In this review, we summarize recent studies of mitotic DSB repair and discuss the mechanisms of suppression of DSB repair during mitosis. DSB repair, which maintains genomic integrity in other phases of the cell cycle, is rather toxic to cells during mitosis, often resulting in chromosome missegregation and aberration. Cells have multiple safeguards to prevent genomic instability during mitosis: inhibition of 53BP1 or BRCA1 localization to DSB sites, which is important to promote non-homologous end joining or homologous recombination, respectively, and also modulation of the non-homologous end joining core complex to inhibit DSB repair. We discuss how DSBs during mitosis are toxic and the multiple safeguard systems that suppress genomic instability.
Keywords: Chromosome segregation; DSB repair; NHEJ; homologous recombination; mitosis.
© 2014 The Authors. Cancer Science published by Wiley Publishing Asia Pty Ltd on behalf of Japanese Cancer Association.
Figures


References
-
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. - PubMed
-
- Bogue MA, Wang C, Zhu C, Roth DB. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

