Computational design of a self-assembling symmetrical β-propeller protein
- PMID: 25288768
- PMCID: PMC4210308
- DOI: 10.1073/pnas.1412768111
Computational design of a self-assembling symmetrical β-propeller protein
Abstract
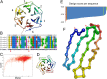
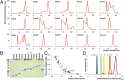
The modular structure of many protein families, such as β-propeller proteins, strongly implies that duplication played an important role in their evolution, leading to highly symmetrical intermediate forms. Previous attempts to create perfectly symmetrical propeller proteins have failed, however. We have therefore developed a new and rapid computational approach to design such proteins. As a test case, we have created a sixfold symmetrical β-propeller protein and experimentally validated the structure using X-ray crystallography. Each blade consists of 42 residues. Proteins carrying 2-10 identical blades were also expressed and purified. Two or three tandem blades assemble to recreate the highly stable sixfold symmetrical architecture, consistent with the duplication and fusion theory. The other proteins produce different monodisperse complexes, up to 42 blades (180 kDa) in size, which self-assemble according to simple symmetry rules. Our procedure is suitable for creating nano-building blocks from different protein templates of desired symmetry.
Keywords: computational protein design; protein crystallography; protein evolution; self-assembly; β-propeller.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Yadid I, Tawfik DS. Reconstruction of functional beta-propeller lectins via homo-oligomeric assembly of shorter fragments. J Mol Biol. 2007;365(1):10–17. - PubMed
-
- Main ER, Jackson SE, Regan L. The folding and design of repeat proteins: Reaching a consensus. Curr Opin Struct Biol. 2003;13(4):482–489. - PubMed
-
- Main ER, Lowe AR, Mochrie SG, Jackson SE, Regan L. A recurring theme in protein engineering: The design, stability and folding of repeat proteins. Curr Opin Struct Biol. 2005;15(4):464–471. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

