A candidate multimodal functional genetic network for thermal adaptation
- PMID: 25289178
- PMCID: PMC4183952
- DOI: 10.7717/peerj.578
A candidate multimodal functional genetic network for thermal adaptation
Abstract
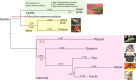
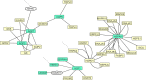
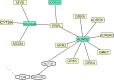
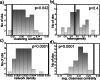
Vertebrate ectotherms such as reptiles provide ideal organisms for the study of adaptation to environmental thermal change. Comparative genomic and exomic studies can recover markers that diverge between warm and cold adapted lineages, but the genes that are functionally related to thermal adaptation may be difficult to identify. We here used a bioinformatics genome-mining approach to predict and identify functions for suitable candidate markers for thermal adaptation in the chicken. We first established a framework of candidate functions for such markers, and then compiled the literature on genes known to adapt to the thermal environment in different lineages of vertebrates. We then identified them in the genomes of human, chicken, and the lizard Anolis carolinensis, and established a functional genetic interaction network in the chicken. Surprisingly, markers initially identified from diverse lineages of vertebrates such as human and fish were all in close functional relationship with each other and more associated than expected by chance. This indicates that the general genetic functional network for thermoregulation and/or thermal adaptation to the environment might be regulated via similar evolutionarily conserved pathways in different vertebrate lineages. We were able to identify seven functions that were statistically overrepresented in this network, corresponding to four of our originally predicted functions plus three unpredicted functions. We describe this network as multimodal: central regulator genes with the function of relaying thermal signal (1), affect genes with different cellular functions, namely (2) lipoprotein metabolism, (3) membrane channels, (4) stress response, (5) response to oxidative stress, (6) muscle contraction and relaxation, and (7) vasodilation, vasoconstriction and regulation of blood pressure. This network constitutes a novel resource for the study of thermal adaptation in the closely related nonavian reptiles and other vertebrate ectotherms.
Keywords: Adaptation; Anolis; Ectotherms; Functional network; Genome mining; Reptiles; Thermal adaptation; Thermoregulation.
Figures


Similar articles
-
Genomic and phenotypic signatures of climate adaptation in an Anolis lizard.Ecol Evol. 2017 Jul 8;7(16):6390-6403. doi: 10.1002/ece3.2985. eCollection 2017 Aug. Ecol Evol. 2017. PMID: 28861242 Free PMC article.
-
Detection of evolutionary conserved and accelerated genomic regions related to adaptation to thermal niches in Anolis lizards.Ecol Evol. 2024 Mar 7;14(3):e11117. doi: 10.1002/ece3.11117. eCollection 2024 Mar. Ecol Evol. 2024. PMID: 38455144 Free PMC article.
-
Functional genomics of abiotic environmental adaptation in lacertid lizards and other vertebrates.J Anim Ecol. 2022 Jun;91(6):1163-1179. doi: 10.1111/1365-2656.13617. Epub 2021 Nov 9. J Anim Ecol. 2022. PMID: 34695234
-
Central nervous regulation of body temperature in vertebrates: comparative aspects.Pharmacol Ther. 1985;30(1):19-30. doi: 10.1016/0163-7258(85)90045-2. Pharmacol Ther. 1985. PMID: 3915819 Review.
-
Thermal adaptation revisited: How conserved are thermal traits of reptiles and amphibians?J Exp Zool A Ecol Integr Physiol. 2021 Jan;335(1):173-194. doi: 10.1002/jez.2414. Epub 2020 Sep 24. J Exp Zool A Ecol Integr Physiol. 2021. PMID: 32970931 Review.
Cited by
-
MALDI-TOF Mass Spectrometry Technology as a Tool for the Rapid Diagnosis of Antimicrobial Resistance in Bacteria.Antibiotics (Basel). 2021 Aug 14;10(8):982. doi: 10.3390/antibiotics10080982. Antibiotics (Basel). 2021. PMID: 34439032 Free PMC article. Review.
-
Genomic and phenotypic signatures of climate adaptation in an Anolis lizard.Ecol Evol. 2017 Jul 8;7(16):6390-6403. doi: 10.1002/ece3.2985. eCollection 2017 Aug. Ecol Evol. 2017. PMID: 28861242 Free PMC article.
-
In silico investigation of uncoupling protein function in avian genomes.Front Vet Sci. 2023 Jan 19;9:1085112. doi: 10.3389/fvets.2022.1085112. eCollection 2022. Front Vet Sci. 2023. PMID: 36744229 Free PMC article.
-
Scans for signatures of selection in Russian cattle breed genomes reveal new candidate genes for environmental adaptation and acclimation.Sci Rep. 2018 Aug 28;8(1):12984. doi: 10.1038/s41598-018-31304-w. Sci Rep. 2018. PMID: 30154520 Free PMC article.
-
Genome-wide landscape of genetic diversity, runs of homozygosity, and runs of heterozygosity in five Alpine and Mediterranean goat breeds.J Anim Sci Biotechnol. 2025 Mar 3;16(1):33. doi: 10.1186/s40104-025-01155-3. J Anim Sci Biotechnol. 2025. PMID: 40025542 Free PMC article.
References
-
- Aggarwal S, Negi S, Jha P, Singh PK, Stobdan T, Pasha MA, Ghosh S, Agrawal A, Indian Genome Variation Consortium. Prasher B, Mukerji M. EGLN1 involvement in high-altitude adaptation revealed through genetic analysis of extreme constitution types defined in Ayurveda. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18961–18966. doi: 10.1073/pnas.1006108107. - DOI - PMC - PubMed
-
- Angilletta MJ. Thermal adaptation: a theoretical and empirical synthesis. New York: Oxford University Press; 2009.
-
- Angilletta MJ. Thermoregulation in animals. In: Angilletta MJ, editor. Oxford bibliographies in physiological ecology of animals. New York: Oxford University Press; 2012.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

