GroopM: an automated tool for the recovery of population genomes from related metagenomes
- PMID: 25289188
- PMCID: PMC4183954
- DOI: 10.7717/peerj.603
GroopM: an automated tool for the recovery of population genomes from related metagenomes
Abstract
Metagenomic binning methods that leverage differential population abundances in microbial communities (differential coverage) are emerging as a complementary approach to conventional composition-based binning. Here we introduce GroopM, an automated binning tool that primarily uses differential coverage to obtain high fidelity population genomes from related metagenomes. We demonstrate the effectiveness of GroopM using synthetic and real-world metagenomes, and show that GroopM produces results comparable with more time consuming, labor-intensive methods.
Keywords: Bioinformatics; Metagenomics; Microbial ecology; Population genome binning.
Figures

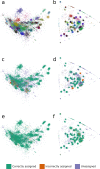

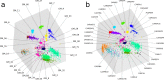
References
-
- Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Loman NJ, Andersson AF, Quince C. CONCOCT: clustering cONtigs on COverage and ComposiTion. 20131312.4038
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

