Functions of FUS/TLS from DNA repair to stress response: implications for ALS
- PMID: 25289647
- PMCID: PMC4189536
- DOI: 10.1177/1759091414544472
Functions of FUS/TLS from DNA repair to stress response: implications for ALS
Abstract
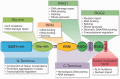
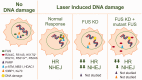
Fused in sarcoma/translocated in liposarcoma (FUS/TLS or FUS) is a multifunctional DNA-/RNA-binding protein that is involved in a variety of cellular functions including transcription, protein translation, RNA splicing, and transport. FUS was initially identified as a fusion oncoprotein, and thus, the early literature focused on the role of FUS in cancer. With the recent discoveries revealing the role of FUS in neurodegenerative diseases, namely amyotrophic lateral sclerosis and frontotemporal lobar degeneration, there has been a renewed interest in elucidating the normal functions of FUS. It is not clear which, if any, endogenous functions of FUS are involved in disease pathogenesis. Here, we review what is currently known regarding the normal functions of FUS with an emphasis on DNA damage repair, RNA processing, and cellular stress response. Further, we discuss how ALS-causing mutations can potentially alter the role of FUS in these pathways, thereby contributing to disease pathogenesis.
Keywords: DNA damage repair; FUS/TLS; RNA processing; amyotrophic lateral sclerosis; stress granules; stress response.
© The Author(s) 2014 Reprints and permissions: sagepub.co.uk/journalsPermissions.nav.
Figures


References
-
- Akhmedov A. T., Bertrand P., Corteggiani E., Lopez B. S. (1995) Characterization of two nuclear mammalian homologous DNA-pairing activities that do not require associated exonuclease activity. Proceedings of the National Academy of Sciences of the United States of America 92: 1729–1733. - PMC - PubMed
-
- Anderson P., Kedersha N. (2008) Stress granules: The Tao of RNA triage. Trends in Biochemical Sciences 33: 141–150. - PubMed
-
- Baechtold H., Kuroda M., Sok J., Ron D., Lopez B. S., Akhmedov A. T. (1999) Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. Journal of Biological Chemistry 274: 34337–34342. - PubMed
-
- Barber S. C., Mead R. J., Shaw P. J. (2006) Oxidative stress in ALS: A mechanism of neurodegeneration and a therapeutic target. Biochimica et Biophysica Acta 1762: 1051–1067. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
