Gut microbial community structure and complications after kidney transplantation: a pilot study
- PMID: 25289916
- PMCID: PMC4189837
- DOI: 10.1097/TP.0000000000000370
Gut microbial community structure and complications after kidney transplantation: a pilot study
Abstract
Background: The gut microbiome plays a role in the regulation of the immune system.
Methods: We prospectively enrolled 26 kidney transplant recipients and collected serial fecal specimens (N=85) during the first three months of transplantation. We characterized bacterial composition by polymerase chain reaction amplification of the 16S rRNA V4-V5 variable region and deep sequencing using the Illumina MiSeq platform.
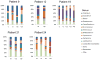
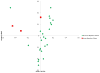
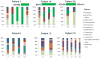
Results: An increase in the relative abundance of Proteobacteria was observed in the posttransplantation specimens compared to pretransplantation specimens (P=0.04, Wilcoxon signed-rank test). In patients with posttransplant diarrhea, the mean(±standard deviation [SD]) Shannon diversity index was lower in those with diarrhea (N=6) than those without diarrhea (N=9) (2.5±0.3 vs. 3.4±0.8; P = 0.02, Wilcoxon rank-sum test). Principal coordinate analysis showed clear separation between the two groups, and linear discriminant analysis effect size (LEfSe) method revealed that Bacteroides, Ruminococcus, Coprococcus, and Dorea were significantly lower in the patients with diarrhea. Principal coordinate analysis also showed clear separation between the acute rejection (AR) group (N=3) and the no AR group (N=23) and the LEfSe method revealed significant differences between the two groups. Fecal abundance of Enterococcus was associated with Enterococcus urinary tract infection (UTI). The median Enterococcus fecal abundance was 24% (range, 8%-95%) in the three patients with Enterococcus UTI compared to 0% in the 23 patients without Enterococcus UTI (interquartile range, 0.00%-0.08%) (P=0.005, Wilcoxon rank-sum test).
Conclusion: Our pilot study identified significant alterations in the gut microbiota after kidney transplantation. Moreover, distinct microbiota structures were observed in allograft recipients with posttransplant diarrhea, AR, and Enterococcus UTI.
Conflict of interest statement
The authors declare no conflict of interest.
Figures











Comment in
-
Gut microbiota: post-transplant complications associated with gut flora.Nat Rev Nephrol. 2014 Nov;10(11):614. doi: 10.1038/nrneph.2014.178. Epub 2014 Sep 23. Nat Rev Nephrol. 2014. PMID: 25247330 No abstract available.
Similar articles
-
Regulation of gut microbiota: a novel pretreatment for complications in patients who have undergone kidney transplantation.Front Cell Infect Microbiol. 2023 Jun 6;13:1169500. doi: 10.3389/fcimb.2023.1169500. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37346031 Free PMC article. Review.
-
Gut microbiota dysbiosis and diarrhea in kidney transplant recipients.Am J Transplant. 2019 Feb;19(2):488-500. doi: 10.1111/ajt.14974. Epub 2018 Jul 21. Am J Transplant. 2019. PMID: 29920927 Free PMC article.
-
The Alteration human of gut microbiota and metabolites before and after renal transplantation.Microb Pathog. 2021 Nov;160:105191. doi: 10.1016/j.micpath.2021.105191. Epub 2021 Sep 24. Microb Pathog. 2021. PMID: 34571151
-
Association of gut microbiota with allograft injury in kidney transplant recipients: a comparative profiling through 16S metagenomics and quantitative PCR.J Med Microbiol. 2024 Nov;73(11). doi: 10.1099/jmm.0.001934. J Med Microbiol. 2024. PMID: 39540836
-
The Gut Microbiota in Kidney Transplantation: A Target for Personalized Therapy?Biology (Basel). 2023 Jan 20;12(2):163. doi: 10.3390/biology12020163. Biology (Basel). 2023. PMID: 36829442 Free PMC article. Review.
Cited by
-
Microbiome in Chronic Kidney Disease.Life (Basel). 2022 Sep 28;12(10):1513. doi: 10.3390/life12101513. Life (Basel). 2022. PMID: 36294946 Free PMC article. Review.
-
Gut microbiome in chronic kidney disease: challenges and opportunities.Transl Res. 2017 Jan;179:24-37. doi: 10.1016/j.trsl.2016.04.007. Epub 2016 Apr 30. Transl Res. 2017. PMID: 27187743 Free PMC article. Review.
-
PREBIOTIC: a study protocol of a randomised controlled trial to assess prebiotic supplementation in kidney transplant recipients for preventing infections and gastrointestinal upset - a feasibility study.Pilot Feasibility Stud. 2023 Jan 16;9(1):11. doi: 10.1186/s40814-023-01236-y. Pilot Feasibility Stud. 2023. PMID: 36647175 Free PMC article.
-
Characteristics and Dysbiosis of the Gut Microbiome in Renal Transplant Recipients.J Clin Med. 2020 Feb 1;9(2):386. doi: 10.3390/jcm9020386. J Clin Med. 2020. PMID: 32024079 Free PMC article.
-
Regulation of gut microbiota: a novel pretreatment for complications in patients who have undergone kidney transplantation.Front Cell Infect Microbiol. 2023 Jun 6;13:1169500. doi: 10.3389/fcimb.2023.1169500. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37346031 Free PMC article. Review.
References
-
- Oh PL, Martinez I, Sun Y, Walter J, Peterson DA, Mercer DF. Characterization of the ileal microbiota in rejecting and nonrejecting recipients of small bowel transplants. Am J Transplant. 2012;12 (3):753. - PubMed
-
- Vaziri ND, Wong J, Pahl M, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83 (2):308. - PubMed
-
- Magurran AE. Biological diversity. Curr Biol. 2005;15 (4):R116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

