Centromere-independent accumulation of cohesin at ectopic heterochromatin sites induces chromosome stretching during anaphase
- PMID: 25290697
- PMCID: PMC4188515
- DOI: 10.1371/journal.pbio.1001962
Centromere-independent accumulation of cohesin at ectopic heterochromatin sites induces chromosome stretching during anaphase
Abstract
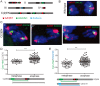
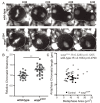
Pericentric heterochromatin, while often considered as "junk" DNA, plays important functions in chromosome biology. It contributes to sister chromatid cohesion, a process mediated by the cohesin complex that ensures proper genome segregation during nuclear division. Long stretches of heterochromatin are almost exclusively placed at centromere-proximal regions but it remains unclear if there is functional (or mechanistic) importance in linking the sites of sister chromatid cohesion to the chromosomal regions that mediate spindle attachment (the centromere). Using engineered chromosomes in Drosophila melanogaster, we demonstrate that cohesin enrichment is dictated by the presence of heterochromatin rather than centromere proximity. This preferential accumulation is caused by an enrichment of the cohesin-loading factor (Nipped-B/NIPBL/Scc2) at dense heterochromatic regions. As a result, chromosome translocations containing ectopic pericentric heterochromatin embedded in euchromatin display additional cohesin-dependent constrictions. These ectopic cohesion sites, placed away from the centromere, disjoin abnormally during anaphase and chromosomes exhibit a significant increase in length during anaphase (termed chromatin stretching). These results provide evidence that long stretches of heterochromatin distant from the centromere, as often found in many cancers, are sufficient to induce abnormal accumulation of cohesin at these sites and thereby compromise the fidelity of chromosome segregation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Topp CN, Dawe RK (2006) Reinterpreting pericentromeric heterochromatin. Curr Opin Plant Biol 9: 647–653. - PubMed
-
- Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45. - PubMed
-
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K (2008) The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301. - PubMed
-
- Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400: 37–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

