Co-ordinated brain and craniofacial development depend upon Patched1/XIAP regulation of cell survival
- PMID: 25292199
- PMCID: PMC4291248
- DOI: 10.1093/hmg/ddu489
Co-ordinated brain and craniofacial development depend upon Patched1/XIAP regulation of cell survival
Abstract
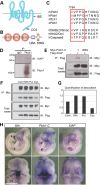
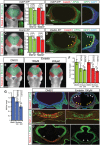
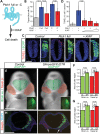
Congenital brain and craniofacial defects often occur together as a consequence of their developmental dependency on common progenitor tissue interactions and signaling pathways during embryogenesis. A classic example of this is perturbation of midline embryo development, and disruption of Hedgehog (Hh) signaling in the pathogenesis of holoprosencephaly. However, our understanding of how Hh signaling governs cell and tissue survival remains incomplete. Patched1 (Ptch1) is a well-known receptor for Hh ligands and Ptch1 overexpression is associated with cell and tissue-specific apoptosis. Here, we demonstrate that the X-linked inhibitory apoptosis protein (XIAP) associates with the C terminus of Ptch1 (Ptch1-C) in primary cilia to inhibit Ptch1-mediated cell death. Consistent with this observation, inhibition of XIAP suppresses cell proliferation, resulting in cell death and pathogenesis of an Hh loss-of-function phenotype. Thus, co-ordinated development of the brain and face is dependent in part upon XIAP mediation of Hh/Ptch1-regulated cell survival and apoptosis during embryogenesis.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Demyer W., Zeman W., Palmer C.G. The face predicts the brain: diagnostic significance of median facial anomalies for holoprosencephaly (arhinencephaly) Pediatrics. 1964;34:256–263. - PubMed
-
- Cohen M.M., Jr Perspectives on holoprosencephaly: part III. Spectra, distinctions, continuities, and discontinuities. Am. J. Med. Genet. 1989;34:271–288. - PubMed
-
- Cohen M.M., Jr, Shiota K. Teratogenesis of holoprosencephaly. Am. J. Med. Genet. 2002;109:1–15. - PubMed
-
- Aoto K., Yayoi S., Higashiyama D., Shiota K., Motoyama J. Fetal ethanol exposure activates protein kinase A and impairs Shh expression in prechordal mesendoderm cells in the pathogenesis of holoprosencephaly. Birth Defects Res. A Clin. Mol. Teratol. 2008;82:224–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

