Messenger RNA degradation in bacterial cells
- PMID: 25292357
- PMCID: PMC4431577
- DOI: 10.1146/annurev-genet-120213-092340
Messenger RNA degradation in bacterial cells
Abstract
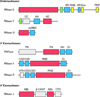
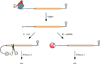
mRNA degradation is an important mechanism for controlling gene expression in bacterial cells. This process involves the orderly action of a battery of cellular endonucleases and exonucleases, some universal and others present only in certain species. These ribonucleases function with the assistance of ancillary enzymes that covalently modify the 5' or 3' end of RNA or unwind base-paired regions. Triggered by initiating events at either the 5' terminus or an internal site, mRNA decay occurs at diverse rates that are transcript specific and governed by RNA sequence and structure, translating ribosomes, and bound sRNAs or proteins. In response to environmental cues, bacteria are able to orchestrate widespread changes in mRNA lifetimes by modulating the concentration or specific activity of cellular ribonucleases or by unmasking the mRNA-degrading activity of cellular toxins.
Keywords: gene regulation; mRNA stability; ribonuclease; sRNA; translation.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

