Functional phylogenomics analysis of bacteria and archaea using consistent genome annotation with UniFam
- PMID: 25293379
- PMCID: PMC4194380
- DOI: 10.1186/s12862-014-0207-y
Functional phylogenomics analysis of bacteria and archaea using consistent genome annotation with UniFam
Abstract
Background: Phylogenetic studies have provided detailed knowledge on the evolutionary mechanisms of genes and species in Bacteria and Archaea. However, the evolution of cellular functions, represented by metabolic pathways and biological processes, has not been systematically characterized. Many clades in the prokaryotic tree of life have now been covered by sequenced genomes in GenBank. This enables a large-scale functional phylogenomics study of many computationally inferred cellular functions across all sequenced prokaryotes.
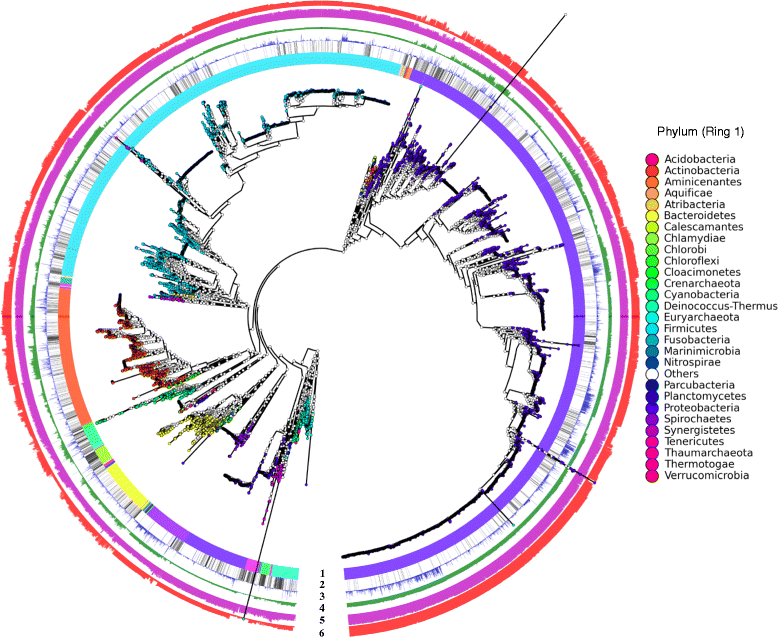
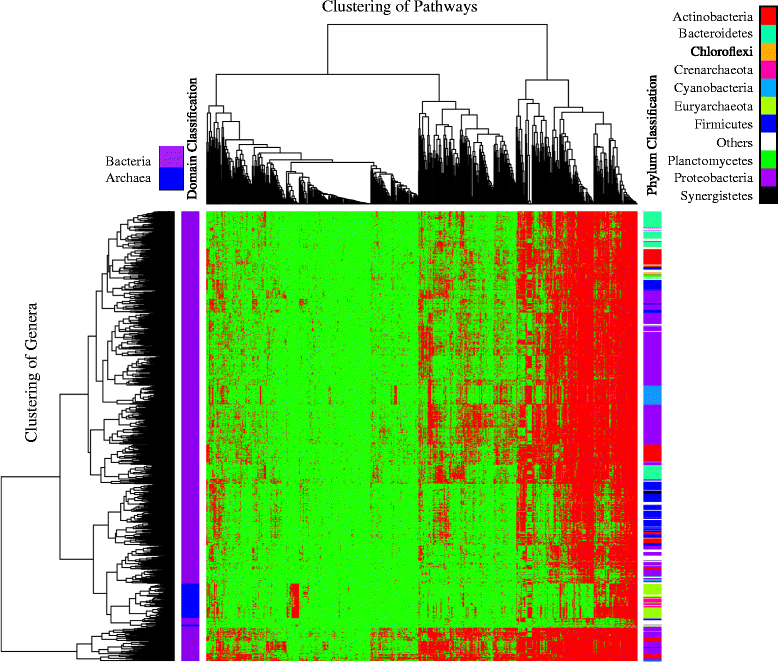
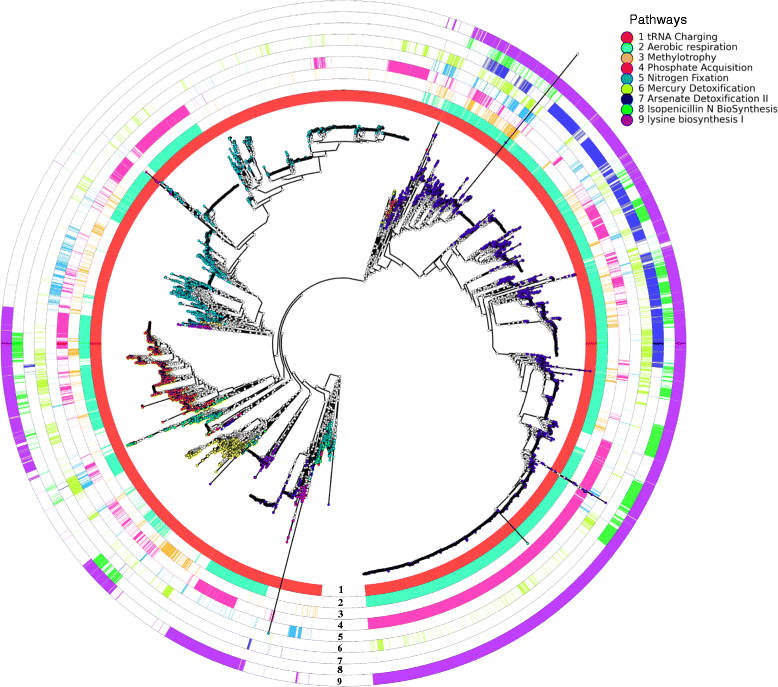
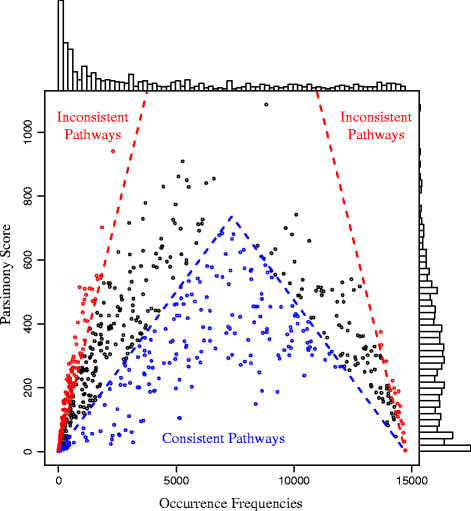
Results: A total of 14,727 GenBank prokaryotic genomes were re-annotated using a new protein family database, UniFam, to obtain consistent functional annotations for accurate comparison. The functional profile of a genome was represented by the biological process Gene Ontology (GO) terms in its annotation. The GO term enrichment analysis differentiated the functional profiles between selected archaeal taxa. 706 prokaryotic metabolic pathways were inferred from these genomes using Pathway Tools and MetaCyc. The consistency between the distribution of metabolic pathways in the genomes and the phylogenetic tree of the genomes was measured using parsimony scores and retention indices. The ancestral functional profiles at the internal nodes of the phylogenetic tree were reconstructed to track the gains and losses of metabolic pathways in evolutionary history.
Conclusions: Our functional phylogenomics analysis shows divergent functional profiles of taxa and clades. Such function-phylogeny correlation stems from a set of clade-specific cellular functions with low parsimony scores. On the other hand, many cellular functions are sparsely dispersed across many clades with high parsimony scores. These different types of cellular functions have distinct evolutionary patterns reconstructed from the prokaryotic tree.
Figures
References
-
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Pujar A, Shearer AG, Travers M, Weerasinghe D, Zhang P, Karp PD. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40(Database issue):D742–D753. doi: 10.1093/nar/gkr1014. - DOI - PMC - PubMed
-
- Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D’Eustachio P, Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39(Database issue):D691–D697. doi: 10.1093/nar/gkq1018. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

