Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits
- PMID: 25294836
- PMCID: PMC4231747
- DOI: 10.1093/nar/gku878
Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits
Abstract
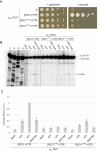
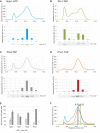
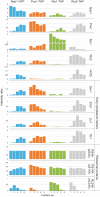
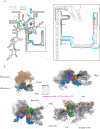
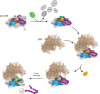
During the last step in 40S ribosome subunit biogenesis, the PIN-domain endonuclease Nob1 cleaves the 20S pre-rRNA at site D, to form the mature 18S rRNAs. Here we report that cleavage occurs in particles that have largely been stripped of previously characterized pre-40S components, but retain the endonuclease Nob1, its binding partner Pno1 (Dim2) and the atypical ATPase Rio1. Within the Rio1-associated pre-40S particles, in vitro pre-rRNA cleavage was strongly stimulated by ATP and required nucleotide binding by Rio1. In vivo binding sites for Rio1, Pno1 and Nob1 were mapped by UV cross-linking in actively growing cells. Nob1 and Pno1 bind overlapping regions within the internal transcribed spacer 1, and both bind directly over cleavage site D. Binding sites for Rio1 were within the core of the 18S rRNA, overlapping tRNA interaction sites and distinct from the related kinase Rio2. Site D cleavage occurs within pre-40S-60S complexes and Rio1-associated particles efficiently assemble into these complexes, whereas Pno1 appeared to be depleted relative to Nob1. We speculate that Rio1-mediated dissociation of Pno1 from cleavage site D is the trigger for final 18S rRNA maturation.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

