Molecular phylogeny and evolutionary history of the Eurasiatic orchid genus Himantoglossum s.l. (Orchidaceae)
- PMID: 25294871
- PMCID: PMC4649687
- DOI: 10.1093/aob/mcu179
Molecular phylogeny and evolutionary history of the Eurasiatic orchid genus Himantoglossum s.l. (Orchidaceae)
Abstract
Background and aims: Lizard orchids of the genus Himantoglossum include many of Eurasia's most spectacular orchids, producing substantial spikes of showy flowers. However, until recently the genus had received only limited, and entirely traditional, systematic study. The aim of the current work was to provide a more robust molecular phylogeny in order to better understand the evolutionary relationships among species of particular conservation concern.
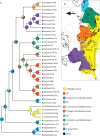
Methods: All putative species of Himantoglossum s.l. were sampled across its geographical range. A large subsample of the 153 populations studied contributed to an initial survey of nuclear ribosomal internal transcribed spacer (nrITS) ribotypes. Smaller subsets were then sequenced for four plastid regions and the first intron of the low-copy-number nuclear gene LEAFY. Rooted using Steveniella as outgroup, phylogenetic trees were generated using parsimony and Bayesian methods from each of the three datasets, supplemented with a ribotype network.
Key results: The resulting trees collectively determined the order of branching of the early divergent taxa as Himantoglossum comperianum > H. robertianum group > H. formosum, events that also involved significant morphological divergence. Relaxed molecular clock dating suggested that these divergences preceded the Pleistocene glaciations (the origin of the H. robertianum group may have coincided with the Messinian salinity crisis) and occurred in Asia Minor and/or the Caucasus. Among more controversial taxa of the H. hircinum-jankae clade, which are only subtly morphologically divergent, topological resolution was poorer and topological incongruence between datasets was consequently greater.
Conclusions: Plastid sequence divergence is broadly consistent with prior, morphologically circumscribed taxa and indicates a division between H. hircinum-adriaticum to the west of the Carpathians and H. jankae-caprinum (plus local endemics) to the east, a distinction also suggested by nrITS ribotypes. LEAFY phylogenies are less congruent with prior taxonomic arrangements and include one likely example of paralogy. Himantoglossum metlesicsianum fully merits its IUCN Endangered status. Potentially significant genetic variation was detected within Steveniella satyrioides, H. robertianum and H. hircinum. However, confident circumscription of the more derived species of Himantoglossum s.s., including local endemics of hybrid origin, must await future morphometric and population-genetic analyses.
Keywords: Dating; Himantoglossum s.l.; LEAFY; Orchidaceae; evolution; hybridization; incomplete lineage sorting; internal transcribed spacer; molecular phylogeny; molecular phylogeography; nrITS; orchid; topological incongruence.
© The Author 2014. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures








References
-
- Acevedo Rodriguez A, Mesa Coello R. 152: Adiciones corologicas de Himantoglossum metlesicsianum (W. P. Teschner) P. Delforge (Orchidaceae): primera cita para la isla de La Palma (Islas Canarias) Botanicá Macaronésica. 2013;28:123–128.
-
- Alibertis A, Alibertis C. Crète (la) n'a pas fini de livrer ses secrets (2) L'Orchidophile. 1989;20:110.
-
- Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29:417–434. - PubMed
-
- Bailey CD, Carr TG, Harris SA, Hughes CE. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Molecular Phylogenetics and Evolution. 2003;29:435–455. - PubMed
-
- Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden. 1995;82:247–277.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

