A crowd-sourcing approach for the construction of species-specific cell signaling networks
- PMID: 25294919
- PMCID: PMC4325542
- DOI: 10.1093/bioinformatics/btu659
A crowd-sourcing approach for the construction of species-specific cell signaling networks
Abstract
Motivation: Animal models are important tools in drug discovery and for understanding human biology in general. However, many drugs that initially show promising results in rodents fail in later stages of clinical trials. Understanding the commonalities and differences between human and rat cell signaling networks can lead to better experimental designs, improved allocation of resources and ultimately better drugs.
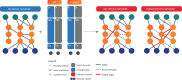
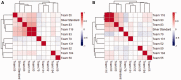
Results: The sbv IMPROVER Species-Specific Network Inference challenge was designed to use the power of the crowds to build two species-specific cell signaling networks given phosphoproteomics, transcriptomics and cytokine data generated from NHBE and NRBE cells exposed to various stimuli. A common literature-inspired reference network with 220 nodes and 501 edges was also provided as prior knowledge from which challenge participants could add or remove edges but not nodes. Such a large network inference challenge not based on synthetic simulations but on real data presented unique difficulties in scoring and interpreting the results. Because any prior knowledge about the networks was already provided to the participants for reference, novel ways for scoring and aggregating the results were developed. Two human and rat consensus networks were obtained by combining all the inferred networks. Further analysis showed that major signaling pathways were conserved between the two species with only isolated components diverging, as in the case of ribosomal S6 kinase RPS6KA1. Overall, the consensus between inferred edges was relatively high with the exception of the downstream targets of transcription factors, which seemed more difficult to predict.
Contact: ebilal@us.ibm.com or gustavo@us.ibm.com.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2014. Published by Oxford University Press.
Figures



References
-
- Calegari VC, et al. Suppressor of cytokine signaling 3 is induced by angiotensin II in heart and isolated cardiomyocytes, and participates in desensitization. Endocrinology. 2003;144:4586–4596. - PubMed
-
- Chen T, et al. Modeling gene expression with differential equations. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 1999;4:29–40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

