Cell cycle control and seed development
- PMID: 25295050
- PMCID: PMC4171995
- DOI: 10.3389/fpls.2014.00493
Cell cycle control and seed development
Abstract
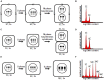
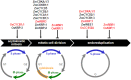
Seed development is a complex process that requires coordinated integration of many genetic, metabolic, and physiological pathways and environmental cues. Different cell cycle types, such as asymmetric cell division, acytokinetic mitosis, mitotic cell division, and endoreduplication, frequently occur in sequential yet overlapping manner during the development of the embryo and the endosperm, seed structures that are both products of double fertilization. Asymmetric cell divisions in the embryo generate polarized daughter cells with different cell fates. While nuclear and cell division cycles play a key role in determining final seed cell numbers, endoreduplication is often associated with processes such as cell enlargement and accumulation of storage metabolites that underlie cell differentiation and growth of the different seed compartments. This review focuses on recent advances in our understanding of different cell cycle mechanisms operating during seed development and their impact on the growth, development, and function of seed tissues. Particularly, the roles of core cell cycle regulators, such as cyclin-dependent-kinases and their inhibitors, the Retinoblastoma-Related/E2F pathway and the proteasome-ubiquitin system, are discussed in the contexts of different cell cycle types that characterize seed development. The contributions of nuclear and cellular proliferative cycles and endoreduplication to cereal endosperm development are also discussed.
Keywords: cell division; cotyledon; cyclin-dependent kinase; embryo; endoreduplication; endosperm; retinoblastoma-related; seed coat.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

