Aptamers as both drugs and drug-carriers
- PMID: 25295268
- PMCID: PMC4177733
- DOI: 10.1155/2014/697923
Aptamers as both drugs and drug-carriers
Abstract
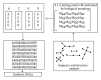
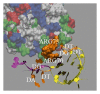
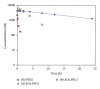
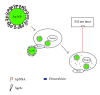
Aptamers are short nucleic acid oligos. They may serve as both drugs and drug-carriers. Their use as diagnostic tools is also evident. They can be generated using various experimental, theoretical, and computational techniques. The systematic evolution of ligands by exponential enrichment which uses iterative screening of nucleic acid libraries is a popular experimental technique. Theory inspired methodology entropy-based seed-and-grow strategy that designs aptamer templates to bind specifically to targets is another one. Aptamers are predicted to be highly useful in producing general drugs and theranostic drugs occasionally for certain diseases like cancer, Alzheimer's disease, and so on. They bind to various targets like lipids, nucleic acids, proteins, small organic compounds, and even entire organisms. Aptamers may also serve as drug-carriers or nanoparticles helping drugs to get released in specific target regions. Due to better target specific physical binding properties aptamers cause less off-target toxicity effects. Therefore, search for aptamer based drugs, drug-carriers, and even diagnostic tools is expanding fast. The biophysical properties in relation to the target specific binding phenomena of aptamers, energetics behind the aptamer transport of drugs, and the consequent biological implications will be discussed. This review will open up avenues leading to novel drug discovery and drug delivery.
Figures






References
-
- Banerjee A, Luduena RF. Kinetics of colchicine binding to purified β-tubulin isotypes from bovine brain. The Journal of Biological Chemistry. 1992;267(19):13335–13339. - PubMed
-
- Haga T, Kurokawa M. Microtubule formation from two components separated by gel filtration of a tubulin preparation. Biochimica et Biophysica Acta: General Subjects. 1975;392(2):335–345. - PubMed
-
- Holmes FA, Kudelka AP, Kavanagh JJ, Huber MH, Ajani JA, Valero V. Current status of clinical trials with paclitaxel and docutaxel. In: Georg GI, Chen TC, Ojima I, Vyas DM, editors. Taxane Anticancer Agents: Basic Science and Current Status. Vol. 583. Washington, DC, USA: American Chemical Society; 1995. pp. 31–57. (ACS Symposium Series).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

