Genetic variation within the histamine pathway among patients with asthma--a pilot study
- PMID: 25295384
- PMCID: PMC4416071
- DOI: 10.3109/02770903.2014.973501
Genetic variation within the histamine pathway among patients with asthma--a pilot study
Abstract
Objective: Histamine is an important mediator in the pathophysiology of asthma. We have previously reported that HRH1 is differentially expressed among those with asthma compared to those without asthma. Single histamine-related genes have also been associated with asthma. We aimed to evaluate known single nucleotide polymorphisms (SNPs) in genes along the histamine biotransformation and response pathway, and determine their association with asthma and HRH1 mRNA expression.
Methods: We enrolled children and adults (n = 93) with/without asthma who met inclusion/exclusion criteria. Genotyping was performed for nine known SNPs in the HDC, HRH1, HRH4, HNMT and ABP1 genes. HRH1 mRNA expression was determined on RNA from buccal tissue. General linear model, Fisher's exact test and Chi-square test were used to determine differences in allele, genotype and haplotype frequency between subjects with and without asthma and differential HRH1 mRNA expression relative to genotype. Statistical significance was determined by p < 0.05.
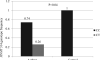
Results: No difference was observed in genotype/allele frequency for the nine SNPs between subjects with and without asthma. The HNMT-1639C/-464C/314C/3'UTRA haplotype was more frequently observed in those without asthma than those with asthma (p = 0.03). We also observed genetic differences relative to race and gender. HNMT 314 genotype CT was more frequent in males with asthma compared to those without asthma (p = 0.04).
Conclusions: Histamine pathway haplotype was associated with a diagnosis of asthma in our cohort but allele and genotype were not. Subgroup evaluations may also be important. Further studies are needed to determine the potential biological/clinical significance of our findings.
Keywords: ABP1; HDC; HNMT; HRH 1; HRH4; mRNA expression; single nucleotide polymorphisms.
Figures
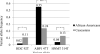



Similar articles
-
Genetic Variation in the Histamine Production, Response, and Degradation Pathway Is Associated with Histamine Pharmacodynamic Response in Children with Asthma.Front Pharmacol. 2017 Jan 4;7:524. doi: 10.3389/fphar.2016.00524. eCollection 2016. Front Pharmacol. 2017. PMID: 28101058 Free PMC article.
-
Genetic Variation along the Histamine Pathway in Children with Allergic versus Nonallergic Asthma.Am J Respir Cell Mol Biol. 2015 Dec;53(6):802-9. doi: 10.1165/rcmb.2014-0493OC. Am J Respir Cell Mol Biol. 2015. PMID: 25909280 Free PMC article.
-
Asthma endophenotypes and polymorphisms in the histamine receptor HRH4 gene.Int Arch Allergy Immunol. 2012;159(2):109-20. doi: 10.1159/000335919. Epub 2012 May 30. Int Arch Allergy Immunol. 2012. PMID: 22653292
-
Histamine pharmacogenomics.Pharmacogenomics. 2009 May;10(5):867-83. doi: 10.2217/pgs.09.26. Pharmacogenomics. 2009. PMID: 19450133 Review.
-
Histamine N-Methyltransferase in the Brain.Int J Mol Sci. 2019 Feb 10;20(3):737. doi: 10.3390/ijms20030737. Int J Mol Sci. 2019. PMID: 30744146 Free PMC article. Review.
Cited by
-
Genetic Mechanisms of Asthma and the Implications for Drug Repositioning.Genes (Basel). 2018 May 3;9(5):237. doi: 10.3390/genes9050237. Genes (Basel). 2018. PMID: 29751569 Free PMC article. Review.
-
Plasma Inflammatory Cytokine IL-4, IL-8, IL-10, and TNF-α Levels Correlate with Pulmonary Function in Patients with Asthma-Chronic Obstructive Pulmonary Disease (COPD) Overlap Syndrome.Med Sci Monit. 2016 Aug 9;22:2800-8. doi: 10.12659/msm.896458. Med Sci Monit. 2016. PMID: 27501772 Free PMC article.
-
Nuclear receptor Nr1d1 alleviates asthma by abating GATA3 gene expression and Th2 cell differentiation.Cell Mol Life Sci. 2022 May 21;79(6):308. doi: 10.1007/s00018-022-04323-0. Cell Mol Life Sci. 2022. PMID: 35596832 Free PMC article.
-
Genetic Variation in the Histamine Production, Response, and Degradation Pathway Is Associated with Histamine Pharmacodynamic Response in Children with Asthma.Front Pharmacol. 2017 Jan 4;7:524. doi: 10.3389/fphar.2016.00524. eCollection 2016. Front Pharmacol. 2017. PMID: 28101058 Free PMC article.
-
Combined impacts of histamine receptor H1 gene polymorphisms and an environmental carcinogen on the susceptibility to and progression of oral squamous cell carcinoma.Aging (Albany NY). 2022 May 19;14(10):4500-4512. doi: 10.18632/aging.204089. Epub 2022 May 19. Aging (Albany NY). 2022. PMID: 35587368 Free PMC article.
References
-
- National Heart, Lung, and Blood Institute Expert Panel Report 3 (EPR3): Guidelines for the Diagnosis and Management of Asthma. 2007 [11/13/2013]. Available from: http://www.nhlbi.nih.gov/guidelines/asthma/asthgdln.htm.
-
- American Lung Association Trends in Asthma Morbidity and Mortality. 2012 Sep; [12/03/2013]. Available from: http://www.lung.org/finding-cures/our-research/trend-reports/asthma-tren....
-
- Garcia-Martin E, Ayuso P, Martinez C, Blanca M, Agundez JA. Histamine pharmacogenomics. Pharmacogenomics. 2009 May;10(5):867–83. PubMed PMID: 19450133. Epub 2009/05/20. eng. - PubMed
-
- Hirota N, Risse PA, Novali M, McGovern T, Al-Alwan L, McCuaig S, Proud D, et al. Histamine may induce airway remodeling through release of epidermal growth factor receptor ligands from bronchial epithelial cells. FASEB J. Apr;26(4):1704–16. PubMed PMID: 22247333. Epub 2012/01/17. eng. - PubMed
-
- Igaz P, Fitzimons CP, Szalai C, Falus A. Histamine genomics in silico: polymorphisms of the human genes involved in the synthesis, action and degradation of histamine. Am J Pharmacogenomics. 2002;2(1):67–72. PubMed PMID: 12083955. Epub 2002/06/27. eng. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
