Computational studies of the effect of the S23D/S24D troponin I mutation on cardiac troponin structural dynamics
- PMID: 25296321
- PMCID: PMC4190606
- DOI: 10.1016/j.bpj.2014.08.008
Computational studies of the effect of the S23D/S24D troponin I mutation on cardiac troponin structural dynamics
Abstract
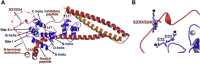
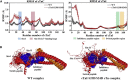
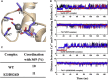
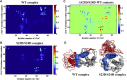
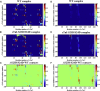
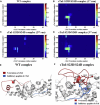
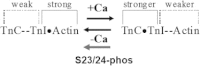
During β-adrenergic stimulation, cardiac troponin I (cTnI) is phosphorylated by protein kinase A (PKA) at sites S23/S24, located at the N-terminus of cTnI. This phosphorylation has been shown to decrease KCa and pCa50, and weaken the cTnC-cTnI (C-I) interaction. We recently reported that phosphorylation results in an increase in the rate of early, slow phase of relaxation (kREL,slow) and a decrease in its duration (tREL,slow), which speeds up the overall relaxation. However, as the N-terminus of cTnI (residues 1-40) has not been resolved in the whole cardiac troponin (cTn) structure, little is known about the molecular-level behavior within the whole cTn complex upon phosphorylation of the S23/S24 residues of cTnI that results in these changes in function. In this study, we built up the cTn complex structure (including residues cTnC 1-161, cTnI 1-172, and cTnT 236-285) with the N-terminus of cTnI. We performed molecular-dynamics (MD) simulations to elucidate the structural basis of PKA phosphorylation-induced changes in cTn structure and Ca(2+) binding. We found that introducing two phosphomimic mutations into sites S23/S24 had no significant effect on the coordinating residues of Ca(2+) binding site II. However, the overall fluctuation of cTn was increased and the C-I interaction was altered relative to the wild-type model. The most significant changes involved interactions with the N-terminus of cTnI. Interestingly, the phosphomimic mutations led to the formation of intrasubunit interactions between the N-terminus and the inhibitory peptide of cTnI. This may result in altered interactions with cTnC and could explain the increased rate and decreased duration of slow-phase relaxation seen in myofibrils.
Copyright © 2014 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Gordon A.M., Homsher E., Regnier M. Regulation of contraction in striated muscle. Physiol. Rev. 2000;80:853–924. - PubMed
-
- Farah C.S., Reinach F.C. The troponin complex and regulation of muscle contraction. FASEB J. 1995;9:755–767. - PubMed
-
- Zhang R., Zhao J., Potter J.D. Cardiac troponin I phosphorylation increases the rate of cardiac muscle relaxation. Circ. Res. 1995;76:1028–1035. - PubMed
-
- Kentish J.C., McCloskey D.T., Solaro R.J. Phosphorylation of troponin I by protein kinase A accelerates relaxation and crossbridge cycle kinetics in mouse ventricular muscle. Circ. Res. 2001;88:1059–1065. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

