Molecular cloning, sequence analysis, and cadmium stress-rated expression changes of BTG1 in freshwater pearl mussel (Hyriopsis schlegelii)
- PMID: 25297078
- PMCID: PMC4790355
- DOI: 10.13918/j.issn.2095-8137.2014.5.389
Molecular cloning, sequence analysis, and cadmium stress-rated expression changes of BTG1 in freshwater pearl mussel (Hyriopsis schlegelii)
Abstract
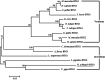
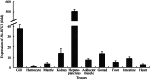
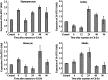
The B cells translocation gene 1 (BTG1) is a member of the BTG/TOB family of anti-proliferative genes, which have recently emerged as important regulators of cell growth and differentiation among verteates. Here, for the first time we cloned the full-length cDNA sequence of Hyriopsis schlegelii (Hs-BTG1), an economically important freshwater shellfish and potential indicator of environmental heavy metal pollution, for the first time. Using rapid amplification of cDNA ends (RACE) together with splicing the EST sequence from a haemocyte cDNA liary, we found that Hs-BTG1 contains a 525 bp open reading frame (ORF) encoding a 174 amino-acid polypeptide, a 306 bp 5' untranslated region (5' UTR), and a 571 bp 3' UTR with a Poly(A) tail as well as a transcription termination signal (AATAAA). Homologue searching against GenBank revealed that Hs-BTG1 was closest to Crassostrea gigas BTG1, sharing 50.57% of protein identities. Hs-BTG1 also shares some typical features of the BTG/TOB family, possessing two well-conserved A and B boxes. Clustering analysis of Hs-BTG1 and other known BTGs showed that Hs-BTG1 was also closely related to BTG1 of C. gigas from the inverteate BTG1 clade. Function prediction via homology modeling showed that both Hs-BTG1 and C. gigas BTG1 share a similar three-dimensional structure with Homo sapiens BTG1. Tissue-specific expression analysis of the Hs-BTG1 via real-time PCR showed that the transcripts were constitutively expressed, with the highest levels in the hepatopancreas and gills, and the lowest in both haemocyte and muscle tissue. Expression levels of Hs-BTG1 in hepatopancreas (2.03-fold), mantle (2.07-fold), kidney (2.2-fold) and haemocyte (2.5-fold) were enhanced by cadmium (Cd²⁺) stress, suggesting that Hs-BTG1 may have played a significant role in H. schlegelii adaptation to adverse environmental conditions.
Keywords: BTG1; Cadmium stress; Gene cloning; Hyriopsis schlegelii; mRNA expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Busson M, Carazo A, Seyer P, Grandemange S, Casas F, Pessemesse L, Rouault JP, Wrutniak-Cabello C, Cabello G. 2005. Coactivation of nuclear receptors and myogenic factors induces the major BTG1 influence on muscle differentiation. Oncogene, 24(10): 1698-710. - PubMed
-
- Corjay MH, Kearney MA, Munzer DA, Diamond SM, Stoltenborg JK. 1998. Antiproliferative gene BTG1 is highly expressed in apoptotic cells in macrophage-rich areas of advanced lesions in Watanabe heritable hyperlipidemic rabbit and human. Laboratory Investigation, 78: 847-58. - PubMed
-
- Duan YF, Liu P, Li JT, Li J, Gao BQ, Chen P. 2013. Cloning and expression analysis of Cathepsin L cDNA of Exopalaemon carinicauda. Zoological Research, 34(1): 39-46.(in Chinese) - PubMed
-
- Fu YJ, Huang FG, Yuan T, Gu JR, Luo GQ, Xu H. 2012. Molecular cloning, characterization and expression analysis of B cell translocation gene 1 in grass carp Ctenopharyngodon idella. Journal of Fish Biology, 80(3): 669-78. - PubMed
-
- Feng Z, Shen H, Du ZQ, Zhu MJ, Fan B, Rothschild MF, Zhao SH, Li CC. 2011. BTG1 as a new candidate gene for muscle growth in pigs: Cloning, expression and association analysis. Journal of Animal Science and Biotechnology, 2(3): 121-30.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
