Dye label interference with RNA modification reveals 5-fluorouridine as non-covalent inhibitor
- PMID: 25300485
- PMCID: PMC4227767
- DOI: 10.1093/nar/gku908
Dye label interference with RNA modification reveals 5-fluorouridine as non-covalent inhibitor
Abstract
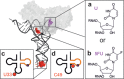
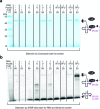
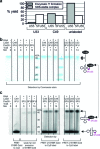
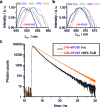
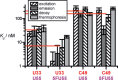
The interest in RNA modification enzymes surges due to their involvement in epigenetic phenomena. Here we present a particularly informative approach to investigate the interaction of dye-labeled RNA with modification enzymes. We investigated pseudouridine (Ψ) synthase TruB interacting with an alleged suicide substrate RNA containing 5-fluorouridine (5FU). A longstanding dogma, stipulating formation of a stable covalent complex was challenged by discrepancies between the time scale of complex formation and enzymatic turnover. Instead of classic mutagenesis, we used differentially positioned fluorescent labels to modulate substrate properties in a range of enzymatic conversion between 6% and 99%. Despite this variegation, formation of SDS-stable complexes occurred instantaneously for all 5FU-substrates. Protein binding was investigated by advanced fluorescence spectroscopy allowing unprecedented simultaneous detection of change in fluorescence lifetime, anisotropy decay, as well as emission and excitation maxima. Determination of Kd values showed that introduction of 5FU into the RNA substrate increased protein affinity by 14× at most. Finally, competition experiments demonstrated reversibility of complex formation for 5FU-RNA. Our results lead us to conclude that the hitherto postulated long-term covalent interaction of TruB with 5FU tRNA is based on the interpretation of artifacts. This is likely true for the entire class of pseudouridine synthases.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Carell T., Brandmayr C., Hienzsch A., Müller M., Pearson D., Reiter V., Thoma I., Thumbs P., Wagner M. Structure and function of noncanonical nucleobases. Angewandte Chemie. 2012;51:7110–7131. - PubMed
-
- Kellner S., Neumann J., Rosenkranz D., Lebedeva S., Ketting R.F., Zischler H., Schneider D., Helm M. Profiling of RNA modifications by multiplexed stable isotope labelling. Chem. Commun. 2014;50:3516–3518. - PubMed
-
- Ramamurthy V., Swann S.L., Paulson J.L., Spedaliere C.J., Mueller E.G. Critical aspartic acid residues in pseudouridine synthases. J. Biol. Chem. 1999;274:22225–22230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

