Profiling the transcription factor regulatory networks of human cell types
- PMID: 25300490
- PMCID: PMC4227771
- DOI: 10.1093/nar/gku923
Profiling the transcription factor regulatory networks of human cell types
Abstract
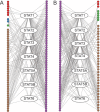
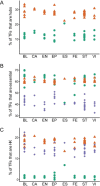
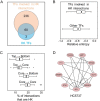
Neph et al. (2012) (Circuitry and dynamics of human transcription factor regulatory networks. Cell, 150: 1274-1286) reported the transcription factor (TF) regulatory networks of 41 human cell types using the DNaseI footprinting technique. This provides a valuable resource for uncovering regulation principles in different human cells. In this paper, the architectures of the 41 regulatory networks and the distributions of housekeeping and specific regulatory interactions are investigated. The TF regulatory networks of different human cell types demonstrate similar global three-layer (top, core and bottom) hierarchical architectures, which are greatly different from the yeast TF regulatory network. However, they have distinguishable local organizations, as suggested by the fact that wiring patterns of only a few TFs are enough to distinguish cell identities. The TF regulatory network of human embryonic stem cells (hESCs) is dense and enriched with interactions that are unseen in the networks of other cell types. The examination of specific regulatory interactions suggests that specific interactions play important roles in hESCs.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Babu M.M., Luscombe N.M., Aravind L., Gerstein M., Teichmann S.A. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004;14:283–291. - PubMed
-
- Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. - PubMed
-
- Davidson E.H. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic Press; 2010.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

