Locus-specific control of DNA resection and suppression of subtelomeric VSG recombination by HAT3 in the African trypanosome
- PMID: 25300492
- PMCID: PMC4227765
- DOI: 10.1093/nar/gku900
Locus-specific control of DNA resection and suppression of subtelomeric VSG recombination by HAT3 in the African trypanosome
Abstract
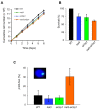
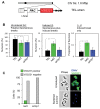
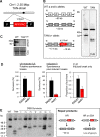
The African trypanosome, Trypanosoma brucei, is a parasitic protozoan that achieves antigenic variation through DNA-repair processes involving Variant Surface Glycoprotein (VSG) gene rearrangements at subtelomeres. Subtelomeric suppression of DNA repair operates in eukaryotes but little is known about these controls in trypanosomes. Here, we identify a trypanosome histone acetyltransferase (HAT3) and a deacetylase (SIR2rp1) required for efficient RAD51-dependent homologous recombination. HAT3 and SIR2rp1 were required for RAD51-focus assembly and disassembly, respectively, at a chromosome-internal locus and a synthetic defect indicated distinct contributions to DNA repair. Although HAT3 promoted chromosome-internal recombination, it suppressed subtelomeric VSG recombination, and these locus-specific effects were mediated through differential production of ssDNA by DNA resection; HAT3 promoted chromosome-internal resection but suppressed subtelomeric resection. Consistent with the resection defect, HAT3 was specifically required for the G2-checkpoint response at a chromosome-internal locus. HAT3 also promoted resection at a second chromosome-internal locus comprising tandem-duplicated genes. We conclude that HAT3 and SIR2rp1 can facilitate temporally distinct steps in DNA repair. HAT3 promotes ssDNA formation and recombination at chromosome-internal sites but has the opposite effect at a subtelomeric VSG. These locus-specific controls reveal compartmentalization of the T. brucei genome in terms of the DNA-damage response and suppression of antigenic variation by HAT3.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Brun R., Blum J., Chappuis F., Burri C. Human African trypanosomiasis. Lancet. 2010;375:148–159. - PubMed
-
- Kostriken R., Strathern J.N., Klar A.J., Hicks J.B., Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983;35:167–174. - PubMed
-
- Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. - PubMed
-
- Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 1985;41:105–114. - PubMed
-
- Pays E. The variant surface glycoprotein as a tool for adaptation in African trypanosomes. Microbes Infect. 2006;8:930–937. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

