Inflammation-driven carcinogenesis is mediated through STING
- PMID: 25300616
- PMCID: PMC4998973
- DOI: 10.1038/ncomms6166
Inflammation-driven carcinogenesis is mediated through STING
Abstract
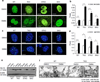
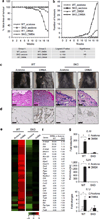
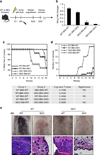
Chronic stimulation of innate immune pathways by microbial agents or damaged tissue is known to promote inflammation-driven tumorigenesis by mechanisms that are not well understood. Here we demonstrate that mutagenic 7,12-dimethylbenz(a)anthracene (DMBA), cisplatin and etoposide induce nuclear DNA leakage into the cytosol that intrinsically activates stimulator of interferon genes (STING)-dependent cytokine production. Inflammatory cytokine levels are subsequently augmented in a STING-dependent extrinsic manner by infiltrating phagocytes purging dying cells. Consequently, STING(-/-) mice, or wild-type mice adoptively transferred with STING(-/-) bone marrow, are almost completely resistant to DMBA-induced skin carcinogenesis compared with their wild-type counterparts. Our data establish a role for STING in the control of cancer, shed significant insight into the causes of inflammation-driven carcinogenesis and may provide a basis for therapeutic strategies to help prevent malignant disease.
Figures

References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

