Frequent migration of introduced cucurbit-infecting begomoviruses among Middle Eastern countries
- PMID: 25300752
- PMCID: PMC4201715
- DOI: 10.1186/1743-422X-11-181
Frequent migration of introduced cucurbit-infecting begomoviruses among Middle Eastern countries
Abstract
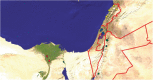
Background: In the early 2000s, two cucurbit-infecting begomoviruses were introduced into the eastern Mediterranean basin: the Old World Squash leaf curl virus (SLCV) and the New World Watermelon chlorotic stunt virus (WmCSV). These viruses have been emerging in parallel over the last decade in Egypt, Israel, Jordan, Lebanon and Palestine.
Methods: We explored this unique situation by assessing the diversity and biogeography of the DNA-A component of SLCV and WmCSV in these five countries.
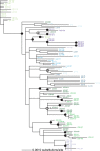
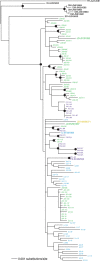
Results: There was fairly low sequence variation in both begomovirus species (SLCV π = 0.0077; WmCSV π = 0.0066). Both viruses may have been introduced only once into the eastern Mediterranean basin, but once established, these viruses readily moved across country boundaries. SLCV has been introduced at least twice into each of all five countries based on the absence of monophyletic clades. Similarly, WmCSV has been introduced multiple times into Jordan, Israel and Palestine.
Conclusions: We predict that uncontrolled movement of whiteflies among countries in this region will continue to cause SLCV and WmCSV migration, preventing strong genetic differentiation of these viruses among these countries.
Figures


References
-
- Antignus Y, Lachman O, Pearlsman M, Omer S, Yunis H, Messika Y, Uko O, Koren A. Squash leaf curl geminivirus – a new illegal immigrant from the western hemisphere and a threat to cucurbit crops in Israel. Phytoparasitica. 2003;31:415.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

