Comparative genomic analysis of genogroup 1 (Wa-like) rotaviruses circulating in the USA, 2006-2009
- PMID: 25301114
- PMCID: PMC4620586
- DOI: 10.1016/j.meegid.2014.09.021
Comparative genomic analysis of genogroup 1 (Wa-like) rotaviruses circulating in the USA, 2006-2009
Abstract
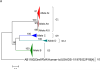
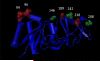
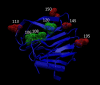
Group A rotaviruses (RVA) are double stranded RNA viruses that are a significant cause of acute pediatric gastroenteritis. Beginning in 2006 and 2008, respectively, two vaccines, Rotarix™ and RotaTeq®, have been approved for use in the USA for prevention of RVA disease. The effects of possible vaccine pressure on currently circulating strains in the USA and their genome constellations are still under investigation. In this study we report 33 complete RVA genomes (ORF regions) collected in multiple cities across USA during 2006-2009, including 8 collected from children with verified receipt of 3 doses of rotavirus vaccine. The strains included 16 G1P[8], 10 G3P[8], and 7 G9P[8]. All 33 strains had a Wa like backbone with the consensus genotype constellation of G(1/3/9)-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1. From maximum likelihood based phylogenetic analyses, we identified 3-7 allelic constellations grouped mostly by respective G types, suggesting a possible allelic segregation based on the VP7 gene of RVA, primarily for the G3 and G9 strains. The vaccine failure strains showed similar grouping for all genes in G9 strains and most genes of G3 strains suggesting that these constellations were necessary to evade vaccine-derived immune protection. Substitutions in the antigenic region of VP7 and VP4 genes were also observed for the vaccine failure strains which could possibly explain how these strains escape vaccine induced immune response. This study helps elucidate how RVA strains are currently evolving in the population post vaccine introduction and supports the need for continued RVA surveillance.
Keywords: Allele; Failure; Rotavirus; VP4; VP7; Vaccine.
Published by Elsevier B.V.
Figures












Similar articles
-
G1P[8] species A rotavirus over 27 years--pre- and post-vaccination eras--in Brazil: full genomic constellation analysis and no evidence for selection pressure by Rotarix® vaccine.Infect Genet Evol. 2015 Mar;30:206-218. doi: 10.1016/j.meegid.2014.12.030. Epub 2015 Jan 3. Infect Genet Evol. 2015. PMID: 25562122
-
G1P[8] Rotavirus in children with severe diarrhea in the post-vaccine introduction era in Brazil: Evidence of reassortments and structural modifications of the antigenic VP7 and VP4 regions.Infect Genet Evol. 2019 Apr;69:255-266. doi: 10.1016/j.meegid.2019.02.009. Epub 2019 Feb 11. Infect Genet Evol. 2019. PMID: 30763774
-
Upsurge and spread of G3 rotaviruses in Eastern India (2014-2016): Full genome analyses reveals heterogeneity within Wa-like genomic constellation.Infect Genet Evol. 2018 Sep;63:158-174. doi: 10.1016/j.meegid.2018.05.026. Epub 2018 May 26. Infect Genet Evol. 2018. PMID: 29842980
-
Circulating rotavirus strains in children with acute gastroenteritis in Iran, 1986 to 2023 and their genetic/antigenic divergence compared to approved vaccines strains (Rotarix, RotaTeq, ROTAVAC, ROTASIIL) before mass vaccination: Clues for vaccination policy makers.Virus Res. 2024 Aug;346:199411. doi: 10.1016/j.virusres.2024.199411. Epub 2024 Jun 3. Virus Res. 2024. PMID: 38823689 Free PMC article. Review.
-
Rotavirus diversity and evolution in the post-vaccine world.Discov Med. 2012 Jan;13(68):85-97. Discov Med. 2012. PMID: 22284787 Free PMC article. Review.
Cited by
-
Genome-Wide Evolutionary Analyses of G1P[8] Strains Isolated Before and After Rotavirus Vaccine Introduction.Genome Biol Evol. 2015 Aug 8;7(9):2473-83. doi: 10.1093/gbe/evv157. Genome Biol Evol. 2015. PMID: 26254487 Free PMC article.
-
Comparative genomic analysis of genogroup 1 and genogroup 2 rotaviruses circulating in seven US cities, 2014-2016.Virus Evol. 2021 Mar 12;7(1):veab023. doi: 10.1093/ve/veab023. eCollection 2021 Jan. Virus Evol. 2021. PMID: 34522389 Free PMC article.
-
Whole-genome characterization of common rotavirus strains circulating in Vellore, India from 2002 to 2017: emergence of non-classical genomic constellations.Gut Pathog. 2023 Sep 20;15(1):44. doi: 10.1186/s13099-023-00569-6. Gut Pathog. 2023. PMID: 37730725 Free PMC article.
-
Rotavirus Strain Trends in United States, 2009-2016: Results from the National Rotavirus Strain Surveillance System (NRSSS).Viruses. 2022 Aug 15;14(8):1775. doi: 10.3390/v14081775. Viruses. 2022. PMID: 36016397 Free PMC article. Review.
-
Amino Acid Substitutions in Positions 385 and 393 of the Hydrophobic Region of VP4 May Be Associated with Rotavirus Attenuation and Cell Culture Adaptation.Viruses. 2020 Apr 7;12(4):408. doi: 10.3390/v12040408. Viruses. 2020. PMID: 32272747 Free PMC article.
References
-
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Systematic biology. 2006;55:539–552. - PubMed
-
- Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, Binka FN, Steele AD, Laserson KF, Ansah NA, Levine MM, Lewis K, Coia ML, Attah-Poku M, Ojwando J, Rivers SB, Victor JC, Nyambane G, Hodgson A, Schodel F, Ciarlet M, Neuzil KM. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;376:606–614. - PubMed
-
- Banyai K, Laszlo B, Duque J, Steele AD, Nelson EA, Gentsch JR, Parashar UD. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30(Suppl 1):A122–130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

