Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids
- PMID: 25302567
- PMCID: PMC4208098
- DOI: 10.1038/ncomms6208
Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids
Abstract
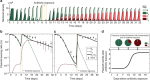
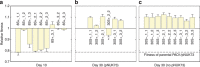
Plasmids are important drivers of bacterial evolution, but it is challenging to understand how plasmids persist over the long term because plasmid carriage is costly. Classical models predict that horizontal transfer is necessary for plasmid persistence, but recent work shows that almost half of plasmids are non-transmissible. Here we use a combination of mathematical modelling and experimental evolution to investigate how a costly, non-transmissible plasmid, pNUK73, can be maintained in populations of Pseudomonas aeruginosa. Compensatory adaptation increases plasmid stability by eliminating the cost of plasmid carriage. However, positive selection for plasmid-encoded antibiotic resistance is required to maintain the plasmid by offsetting reductions in plasmid frequency due to segregational loss. Crucially, we show that compensatory adaptation and positive selection reinforce each other's effects. Our study provides a new understanding of how plasmids persist in bacterial populations, and it helps to explain why resistance can be maintained after antibiotic use is stopped.
Figures




References
-
- Summers D. K. The Biology of Plasmids Blackwell Science Ltd (1996).
-
- Gogarten J. P. & Townsend J. P. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3, 679–687 (2005). - PubMed
-
- Alekshun M. N. & Levy S. B. Molecular mechanisms of antibacterial multidrug resistance. Cell 128, 1037–1050 (2007). - PubMed
-
- eCDC. European Center for Disease Prevention and Control. Annual Epidemiological Report 2012. http://www.ecdc.europa.eu/en/publications/surveillance_reports/Pages/ind... (2013).
-
- CDC. Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States. http://www.cdc.gov/drugresistance/threat-report-2013/ (2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

