Cellular differences in protein synthesis regulate tissue homeostasis
- PMID: 25303523
- PMCID: PMC4222182
- DOI: 10.1016/j.cell.2014.09.016
Cellular differences in protein synthesis regulate tissue homeostasis
Abstract
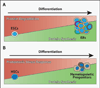
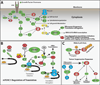
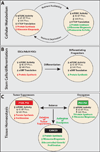
Although sometimes considered a "house-keeping" function, multiple aspects of protein synthesis are regulated differently among somatic cells, including stem cells, and can be modulated in a cell-type-specific manner. These differences are required to establish and maintain differences in cell identity, cell function, tissue homeostasis, and tumor suppression.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
References
-
- Abkowitz JL, Sabo KM, Nakamoto B, Blau CA, Martin FH, Zsebo KM, Papayannopoulou T. Diamond-blackfan anemia: in vitro response of erythroid progenitors to the ligand for c-kit. Blood. 1991;78:2198–2202. - PubMed
-
- Armistead J, Triggs-Raine B. Diverse diseases from a ubiquitous process: The ribosomopathy paradox. FEBS Letters. 2014;588:1491–1500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

