Systems level mapping of metabolic complexity in Mycobacterium tuberculosis to identify high-value drug targets
- PMID: 25304862
- PMCID: PMC4201925
- DOI: 10.1186/s12967-014-0263-5
Systems level mapping of metabolic complexity in Mycobacterium tuberculosis to identify high-value drug targets
Abstract
Background: The effectiveness of current therapeutic regimens for Mycobacterium tuberculosis (Mtb) is diminished by the need for prolonged therapy and the rise of drug resistant/tolerant strains. This global health threat, despite decades of basic research and a wealth of legacy knowledge, is due to a lack of systems level understanding that can innovate the process of fast acting and high efficacy drug discovery.
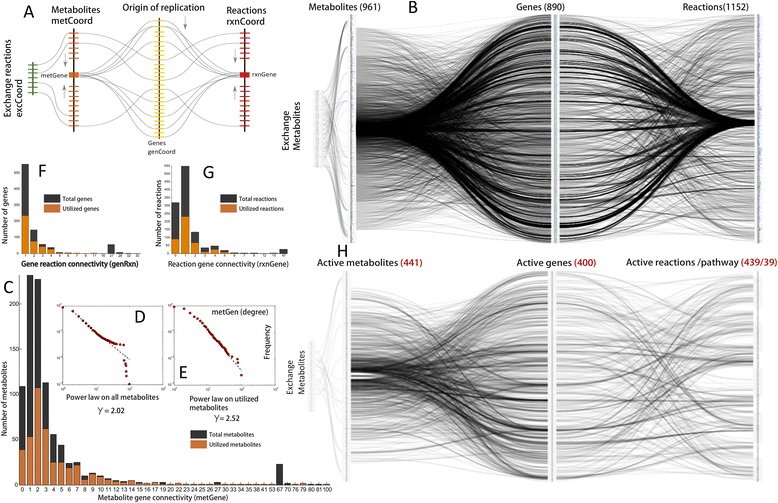
Methods: The enhanced functional annotations of the Mtb genome, which were previously obtained through a crowd sourcing approach was used to reconstruct the metabolic network of Mtb in a bottom up manner. We represent this information by developing a novel Systems Biology Spindle Map of Metabolism (SBSM) and comprehend its static and dynamic structure using various computational approaches based on simulation and design.
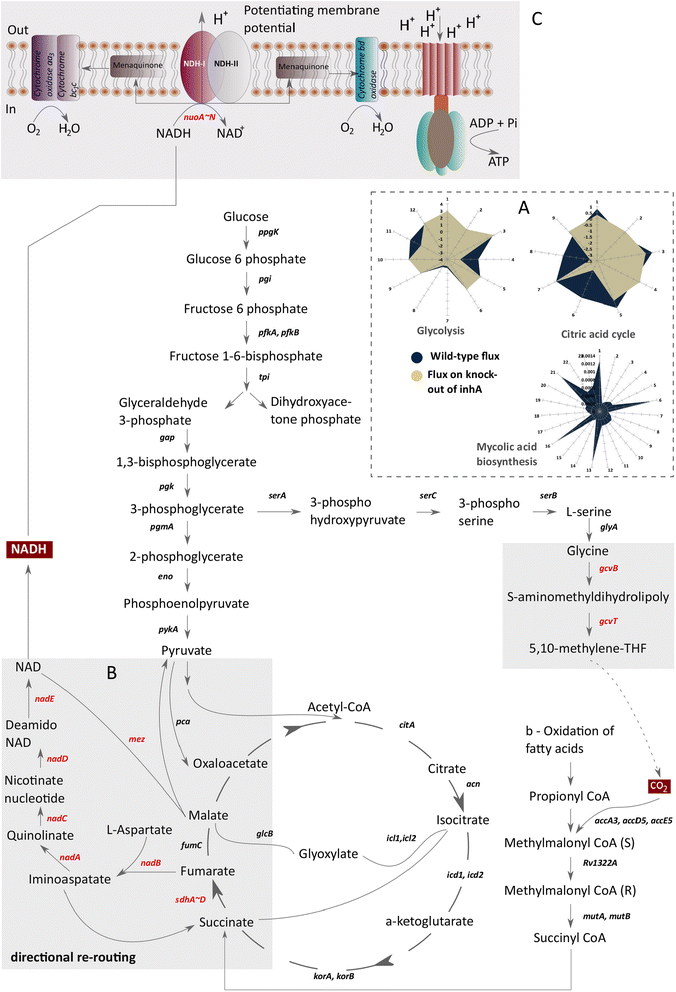
Results: The reconstructed metabolism of Mtb encompasses 961 metabolites, involved in 1152 reactions catalyzed by 890 protein coding genes, organized into 50 pathways. By accounting for static and dynamic analysis of SBSM in Mtb we identified various critical proteins required for the growth and survival of bacteria. Further, we assessed the potential of these proteins as putative drug targets that are fast acting and less toxic. Further, we formulate a novel concept of metabolic persister genes (MPGs) and compared our predictions with published in vitro and in vivo experimental evidence. Through such analyses, we report for the first time that de novo biosynthesis of NAD may give rise to bacterial persistence in Mtb under conditions of metabolic stress induced by conventional anti-tuberculosis therapy. We propose such MPG's as potential combination of drug targets for existing antibiotics that can improve their efficacy and efficiency for drug tolerant bacteria.
Conclusion: The systems level framework formulated by us to identify potential non-toxic drug targets and strategies to circumvent the issue of bacterial persistence can substantially aid in the process of TB drug discovery and translational research.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

