Impact of whole-genome amplification on the reliability of pre-transfer cattle embryo breeding value estimates
- PMID: 25305778
- PMCID: PMC4201692
- DOI: 10.1186/1471-2164-15-889
Impact of whole-genome amplification on the reliability of pre-transfer cattle embryo breeding value estimates
Abstract
Background: Genome-wide profiling of single-nucleotide polymorphisms is receiving increasing attention as a method of pre-implantation genetic diagnosis in humans and of commercial genotyping of pre-transfer embryos in cattle. However, the very small quantity of genomic DNA in biopsy material from early embryos poses daunting technical challenges. A reliable whole-genome amplification (WGA) procedure would greatly facilitate the procedure.
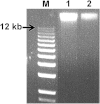
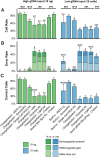
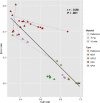
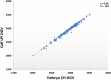
Results: Several PCR-based and non-PCR based WGA technologies, namely multiple displacement amplification, quasi-random primed library synthesis followed by PCR, ligation-mediated PCR, and single-primer isothermal amplification were tested in combination with different DNA extractions protocols for various quantities of genomic DNA inputs. The efficiency of each method was evaluated by comparing the genotypes obtained from 15 cultured cells (representative of an embryonic biopsy) to unamplified reference gDNA. The gDNA input, gDNA extraction method and amplification technology were all found to be critical for successful genome-wide genotyping. The selected WGA platform was then tested on embryo biopsies (n = 226), comparing their results to that of biopsies collected after birth. Although WGA inevitably leads to a random loss of information and to the introduction of erroneous genotypes, following genomic imputation the resulting genetic index of both sources of DNA were highly correlated (r = 0.99, P<0.001).
Conclusion: It is possible to generate high-quality DNA in sufficient quantities for successful genome-wide genotyping starting from an early embryo biopsy. However, imputation from parental and population genotypes is a requirement for completing and correcting genotypic data. Judicious selection of the WGA platform, careful handling of the samples and genomic imputation together, make it possible to perform extremely reliable genomic evaluations for pre-transfer embryos.
Figures



References
-
- Verlinsky Y, Ginsberg N, Lifchez A, Valle J, Moise J, Strom CM. Analysis of the first polar body: preconception genetic diagnosis. Hum Reprod. 1990;5(7):826–829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

