Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7
- PMID: 25306109
- PMCID: PMC4255225
- DOI: 10.1038/nsmb.2902
Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7
Erratum in
-
Corrigendum: Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7.Nat Struct Mol Biol. 2015 Mar;22(3):272. doi: 10.1038/nsmb0315-272b. Nat Struct Mol Biol. 2015. PMID: 25736092 No abstract available.
Abstract
What causes the tissue-specific pathology of diseases resulting from mutations in housekeeping genes? Specifically, in spinocerebellar ataxia type 7 (SCA7), a neurodegenerative disorder caused by a CAG-repeat expansion in ATXN7 (which encodes an essential component of the mammalian transcription coactivation complex, STAGA), the factors underlying the characteristic progressive cerebellar and retinal degeneration in patients were unknown. We found that STAGA is required for the transcription initiation of miR-124, which in turn mediates the post-transcriptional cross-talk between lnc-SCA7, a conserved long noncoding RNA, and ATXN7 mRNA. In SCA7, mutations in ATXN7 disrupt these regulatory interactions and result in a neuron-specific increase in ATXN7 expression. Strikingly, in mice this increase is most prominent in the SCA7 disease-relevant tissues, namely the retina and cerebellum. Our results illustrate how noncoding RNA-mediated feedback regulation of a ubiquitously expressed housekeeping gene may contribute to specific neurodegeneration.
Figures

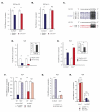
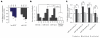

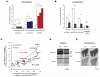
References
-
- Gouw LG, et al. Analysis of the dynamic mutation in the SCA7 gene shows marked parental effects on CAG repeat transmission. Human molecular genetics. 1998;7:525–532. doi: 10.1093/Hmg/7.3.525. - PubMed
-
- David G, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet. 1997;17:65–70. doi: 10.1038/Ng0997-65. - PubMed
-
- Aleman TS, et al. Retinal degeneration associated with spinocerebellar ataxia type 7 (SCA7) Invest Ophthalmol Vis Sci. 2000;41:S175–S175.
-
- Holmberg M, et al. Spinocerebellar ataxia type 7 (SCA7): a neurodegenerative disorder with neuronal intranuclear inclusions. Human molecular genetics. 1998;7:913–918. doi: 10.1093/Hmg/7.5.913. - PubMed
-
- Helmlinger D, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Human molecular genetics. 2004;13:1257–1265. doi: 10.1093/Hmg/Ddh139. - PubMed
METHODS REFERENCES
-
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic acids research. 2008;36:D149–153. microRNA.org. doi: 10.1093/nar/gkm995. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

