Computational approaches for modeling GPCR dimerization
- PMID: 25307013
- PMCID: PMC4237661
- DOI: 10.2174/1389201015666141013102515
Computational approaches for modeling GPCR dimerization
Abstract
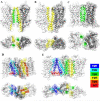
Growing experimental evidences suggest that dimerization and oligomerization are important for G Protein- Coupled Receptors (GPCRs) function. The detailed structural information of dimeric/oligomeric GPCRs would be very important to understand their function. Although it is encouraging that recently several experimental GPCR structures in oligomeric forms have appeared, experimental determination of GPCR structures in oligomeric forms is still a big challenge, especially in mimicking the membrane environment. Therefore, development of computational approaches to predict dimerization of GPCRs will be highly valuable. In this review, we summarize computational approaches that have been developed and used for modeling of GPCR dimerization. In addition, we introduce a novel two-dimensional Brownian Dynamics based protein docking approach, which we have recently adapted, for GPCR dimer prediction.
Figures
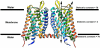


References
-
- Ellis C. The state of GPCR research in 2004. Nat. Rev. Drug Discov. 2004;3(7):575, 577–575, 626. - PubMed
-
- Muller G. Towards 3D structures of G protein-coupled receptors: a multidisciplinary approach. Curr. Med. Chem. 2000;7(9):861–888. - PubMed
-
- Pin JP, Neubig R, Bouvier M, Devi L, Filizola M, Javitch JA, Lohse MJ, Milligan G, Palczewski K, Parmentier M, Spedding M. International Union of Basic and Clinical Pharmacology. LXVII. Recommendations for the recognition and nomenclature of G protein-coupled receptor heteromultimers. Pharmacol. Rev. 2007;59(1):5–13. - PubMed
-
- Filizola M. Advances in Experimental Medicine and Biology: G Protein-Coupled Receptors - Modeling and Simulation, Part II GPCRs in Motion: Insights from Simulations. 2014;796:37–125.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

