A protein-tagging system for signal amplification in gene expression and fluorescence imaging
- PMID: 25307933
- PMCID: PMC4252608
- DOI: 10.1016/j.cell.2014.09.039
A protein-tagging system for signal amplification in gene expression and fluorescence imaging
Abstract
Signals in many biological processes can be amplified by recruiting multiple copies of regulatory proteins to a site of action. Harnessing this principle, we have developed a protein scaffold, a repeating peptide array termed SunTag, which can recruit multiple copies of an antibody-fusion protein. We show that the SunTag can recruit up to 24 copies of GFP, thereby enabling long-term imaging of single protein molecules in living cells. We also use the SunTag to create a potent synthetic transcription factor by recruiting multiple copies of a transcriptional activation domain to a nuclease-deficient CRISPR/Cas9 protein and demonstrate strong activation of endogenous gene expression and re-engineered cell behavior with this system. Thus, the SunTag provides a versatile platform for multimerizing proteins on a target protein scaffold and is likely to have many applications in imaging and controlling biological outputs.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

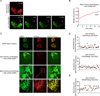
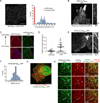

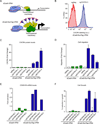
References
-
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nature reviews Molecular cell biology. 2008;9:309–322. - PubMed
-
- Berger C, Weber-Bornhauser S, Eggenberger J, Hanes J, Pluckthun A, Bosshard HR. Antigen recognition by conformational selection. FEBS letters. 1999;450:149–153. - PubMed
-
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Molecular cell. 1998;2:437–445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 CA168370/CA/NCI NIH HHS/United States
- DA036858/DA/NIDA NIH HHS/United States
- P50 GM081879/GM/NIGMS NIH HHS/United States
- DP5 OD017887/OD/NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM38499/GM/NIGMS NIH HHS/United States
- R01 GM038499/GM/NIGMS NIH HHS/United States
- GM081879/GM/NIGMS NIH HHS/United States
- GM102706/GM/NIGMS NIH HHS/United States
- P01 AG010770/AG/NIA NIH HHS/United States
- CA168370/CA/NCI NIH HHS/United States
- P50 GM102706/GM/NIGMS NIH HHS/United States
- R01 DA036858/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

