Central topography of cranial motor nuclei controlled by differential cadherin expression
- PMID: 25308074
- PMCID: PMC4228048
- DOI: 10.1016/j.cub.2014.08.067
Central topography of cranial motor nuclei controlled by differential cadherin expression
Abstract
Neuronal nuclei are prominent, evolutionarily conserved features of vertebrate central nervous system (CNS) organization. Nuclei are clusters of soma of functionally related neurons and are located in highly stereotyped positions. Establishment of this CNS topography is critical to neural circuit assembly. However, little is known of either the cellular or molecular mechanisms that drive nucleus formation during development, a process termed nucleogenesis. Brainstem motor neurons, which contribute axons to distinct cranial nerves and whose functions are essential to vertebrate survival, are organized exclusively as nuclei. Cranial motor nuclei are composed of two main classes, termed branchiomotor/visceromotor and somatomotor. Each of these classes innervates evolutionarily distinct structures, for example, the branchial arches and eyes, respectively. Additionally, each class is generated by distinct progenitor cell populations and is defined by differential transcription factor expression; for example, Hb9 distinguishes somatomotor from branchiomotor neurons. We characterized the time course of cranial motornucleogenesis, finding that despite differences in cellular origin, segregation of branchiomotor and somatomotor nuclei occurs actively, passing through a phase of each being intermingled. We also found that differential expression of cadherin cell adhesion family members uniquely defines each motor nucleus. We show that cadherin expression is critical to nucleogenesis as its perturbation degrades nucleus topography predictably.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
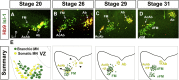


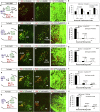
Comment in
-
Neuronal organization: unsticking the cadherin code.Curr Biol. 2014 Dec 1;24(23):R1127-9. doi: 10.1016/j.cub.2014.10.034. Epub 2014 Dec 1. Curr Biol. 2014. PMID: 25465332
References
-
- Cajal S.R.Y. Volume 1. Oxford University Press; New York: 1995. p. 14. (Histology of the Nervous System of Man and Vertebrates).
-
- Agarwala S., Ragsdale C.W. A role for midbrain arcs in nucleogenesis. Development. 2002;129:5779–5788. - PubMed
-
- Kawauchi D., Taniguchi H., Watanabe H., Saito T., Murakami F. Direct visualization of nucleogenesis by precerebellar neurons: involvement of ventricle-directed, radial fibre-associated migration. Development. 2006;133:1113–1123. - PubMed
-
- Nishida K., Nakayama K., Yoshimura S., Murakami F. Role of Neph2 in pontine nuclei formation in the developing hindbrain. Mol. Cell. Neurosci. 2011;46:662–670. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

