The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation
- PMID: 25308709
- PMCID: PMC4664186
- DOI: 10.1111/cmi.12380
The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation
Abstract
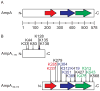
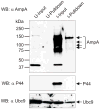
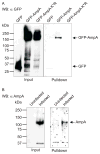
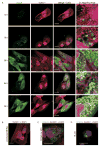
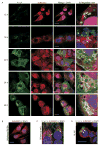
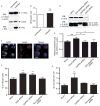
SUMOylation, the covalent attachment of a member of the small ubiquitin-like modifier (SUMO) family of proteins to lysines in target substrates, is an essential post-translational modification in eukaryotes. Microbial manipulation of SUMOylation recently emerged as a key virulence strategy for viruses and facultative intracellular bacteria, the latter of which have only been shown to deploy effectors that negatively regulate SUMOylation. Here, we demonstrate that the obligate intracellular bacterium, Anaplasma phagocytophilum, utilizes an effector, AmpA (A. phagocytophilum post-translationally modified protein A) that becomes SUMOylated in host cells and this is important for the pathogen's survival. We previously discovered that AmpA (formerly APH1387) localizes to the A. phagocytophilum-occupied vacuolar membrane (AVM). Algorithmic prediction analyses denoted AmpA as a candidate for SUMOylation. We verified this phenomenon using a SUMO affinity matrix to precipitate both native AmpA and ectopically expressed green fluorescent protein (GFP)-tagged AmpA. SUMOylation of AmpA was lysine dependent, as SUMO affinity beads failed to precipitate a GFP-AmpA protein when its lysine residues were substituted with arginine. Ectopically expressed and endogenous AmpA were poly-SUMOylated, which was consistent with the observation that AmpA colocalizes with SUMO2/3 at the AVM. Only late during the infection cycle did AmpA colocalize with SUMO1, which terminally caps poly-SUMO2/3 chains. AmpA was also detected in the cytosol of infected host cells, further supporting its secretion and likely participation in interactions that aid pathogen survival. Indeed, whereas siRNA-mediated knockdown of Ubc9 - a necessary enzyme for SUMOylation - slightly bolstered A. phagocytophilum infection, pharmacologically inhibiting SUMOylation in infected cells significantly reduced the bacterial load. Ectopically expressed GFP-AmpA served as a competitive agonist against native AmpA in infected cells, while lysine-deficient GFP-AmpA was less effective, implying that modification of AmpA lysines is important for infection. Collectively, these data show that AmpA becomes directly SUMOylated during infection, representing a novel tactic for A. phagocytophilum survival.
© 2014 John Wiley & Sons Ltd.
Figures


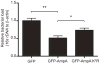
References
-
- Carlyon JA. Establishing intracellular infection: modulation of host cell functions (Anaplasmataceae) In: Palmer GH, Azad A, editors. Intracellular Pathogens II: Rickettsiales. Washington, D. C: ASM Press; 2012.
-
- Carlyon JA, Chan WT, Galan J, Roos D, Fikrig E. Repression of rac2 mRNA expression by Anaplasma phagocytophila is essential to the inhibition of superoxide production and bacterial proliferation. J Immunol. 2002;169:7009–7018. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

