Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry
- PMID: 25309011
- PMCID: PMC4187118
- DOI: 10.1016/j.trac.2014.04.017
Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry
Abstract
Liquid chromatography-mass spectrometry (LC-MS)-based lipidomics has been a subject of dramatic developments over the past decade. This review focuses on state of the art in LC-MS-based lipidomics, covering all the steps of global lipidomic profiling. On the basis of review of 185 original papers and application notes, we can conclude that typical LC-MS-based lipidomics methods involve: (1) extraction using chloroform/MeOH or MTBE/MeOH protocols, both with addition of internal standards covering each lipid class; (2) separation of lipids using short microbore columns with sub-2-μm or 2.6-2.8-μm (fused-core) particle size with C18 or C8 sorbent with analysis time <30 min; (3) electrospray ionization in positive- and negative-ion modes with full spectra acquisition using high-resolution MS with capability to MS/MS. Phospholipids (phosphatidylcholines, phosphatidylethanolamines, phosphatidylinositols, phosphatidylserines, phosphatidylglycerols) followed by sphingomyelins, di- and tri-acylglycerols, and ceramides were the most frequently targeted lipid species.
Keywords: Acylglycerol; Biological system; Comprehensive analysis; Extraction method; Global lipidomic profiling; LC-MS; Lipidomics; Liquid chromatography-mass spectrometry; Metabolomics; Phospholipid.
Figures





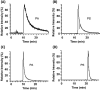
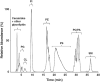
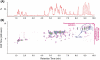
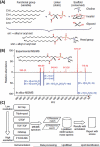
References
-
- Han XL, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44:1071–1079. - PubMed
-
- Hou W, Zhou H, Elisma F, Bennett SA, Figeys D. Technological developments in lipidomics. Brief. Funct. Genomic Proteomic. 2008;7:395–409. - PubMed
-
- Loizides-Mangold U. On the future of mass-spectrometry-based lipidomics. FEBS J. 2013;280:2817–2829. - PubMed
-
- Wenk MR. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005;4:594–610. - PubMed
-
- Li M, Zhou ZG, Nie HG, Bai Y, Liu HW. Recent advances of chromatography and mass spectrometry in lipidomics. Anal. Bioanal. Chem. 2011;399:243–249. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
