An abundant ' Candidatus Phytoplasma solani' tuf b strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus
- PMID: 25309042
- PMCID: PMC4188982
- DOI: 10.1007/s10658-014-0455-0
An abundant ' Candidatus Phytoplasma solani' tuf b strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus
Abstract
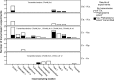
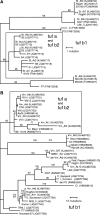
Bois noir (BN) associated with 'Candidatus Phytoplasma solani' (Stolbur) is regularly found in Austrian vine growing regions. Investigations between 2003 and 2008 indicated sporadic presence of the confirmed disease vector Hyalesthes obsoletus and frequent infections of bindweed and grapevine. Infections of nettles were rare. In contrast present investigations revealed a mass occurrence of H. obsoletus almost exclusively on stinging nettle. The high population densities of H. obsoletus on Urtica dioica were accompanied by frequent occurrence of 'Ca. P. solani' in nettles and planthoppers. Sequence analysis of the molecular markers secY, stamp, tuf and vmp1 of stolbur revealed a single genotype named CPsM4_At1 in stinging nettles and more than 64 and 90 % abundance in grapevine and H. obsoletus, respectively. Interestingly, this genotype showed tuf b type restriction pattern previously attributed to bindweed associated 'Ca. P. solani' strains, but a different sequence assigned as tuf b2 compared to reference tuf b strains. All other marker genes of CPsM4_At1 clustered with tuf a and nettle derived genotypes verifying distinct nettle phytoplasma genotypes. Transmission experiments with H. obsoletus and Anaceratagallia ribauti resulted in successful transmission of five different strains including the major genotype to Catharanthus roseus and in transmission of the major genotype to U. dioica. Altogether, five nettle and nine bindweed associated genotypes were described. Bindweed types were verified in 34 % of grapevine samples, in few positive Reptalus panzeri, rarely in bindweeds and occasionally in Catharanthus roseus infected by H. obsoletus or A. ribauti. 'Candidatus Phytoplasma convolvuli' (bindweed yellows) was ascertained in nettle and bindweed samples.
Keywords: Bois noir; Stamp; Stolbur; secY; vmp1; ‘Candidatus phytoplasma convolvuli’.
Figures





References
-
- Bertaccini A, Duduk B. Phytoplasma and phytoplasma diseases: a review of recent research. Phytopathologia Mediterranea. 2009;48:355–378.
-
- COST action FA 0807, 2014. phytoplasma diseases and vectors in europe and surroundings. http://www.costphytoplasma.ipwgnet.org/InsectVectors.htm. Accessed 13 February 2014.
-
- Cvrković T, Jović J, Mitrović M, Krstić O, Toševski I. Experimental and molecular evidence of Reptalus panzeri as a natural vector of bois noir. Plant Pathology. 2014;63:42–53. doi: 10.1111/ppa.12080. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
