Computational Insights into Substrate and Site Specificities, Catalytic Mechanism, and Protonation States of the Catalytic Asp Dyad of β -Secretase
- PMID: 25309776
- PMCID: PMC4189502
- DOI: 10.1155/2014/598728
Computational Insights into Substrate and Site Specificities, Catalytic Mechanism, and Protonation States of the Catalytic Asp Dyad of β -Secretase
Abstract
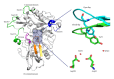
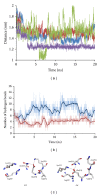
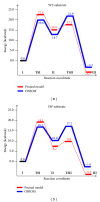
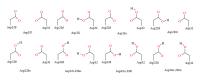
In this review, information regarding substrate and site specificities, catalytic mechanism, and protonation states of the catalytic Asp dyad of β-secretase (BACE1) derived from computational studies has been discussed. BACE1 catalyzes the rate-limiting step in the generation of Alzheimer amyloid beta peptide through the proteolytic cleavage of the amyloid precursor protein. Due to its biological functioning, this enzyme has been considered as one of the most important targets for finding the cure for Alzheimer's disease. Molecular dynamics (MD) simulations suggested that structural differences in the key regions (inserts A, D, and F and the 10s loop) of the enzyme are responsible for the observed difference in its activities towards the WT- and SW-substrates. The modifications in the flap, third strand, and insert F regions were found to be involved in the alteration in the site specificity of the glycosylphosphatidylinositol bound form of BACE1. Our QM and QM/MM calculations suggested that BACE1 hydrolyzed the SW-substrate more efficiently than the WT-substrate and that cleavage of the peptide bond occurred in the rate-determining step. The results from molecular docking studies showed that the information concerning a single protonation state of the Asp dyad is not enough to run an in silico screening campaign.
Figures


References
-
- Suo Z, Humphrey J, Kundtz A, et al. Soluble Alzheimers β-amyloid constricts the cerebral vasculature in vivo. Neuroscience Letters. 1998;257(2):77–80. - PubMed
-
- Pillot T, Drouet B, Queillé S, et al. The nonfibrillar amyloid β-peptide induces apoptotic neuronal cell death: involvement of its C-terminal fusogenic domain. Journal of Neurochemistry. 1999;73(4):1626–1634. - PubMed
-
- Northrop DB. Follow the protons: a low-barrier hydrogen bond unifies the mechanisms of the aspartic proteases. Accounts of Chemical Research. 2001;34(10):790–797. - PubMed
-
- Davies DR. The structure and function of the aspartic proteinases. Annual Review of Biophysics and Biophysical Chemistry. 1990;19:189–215. - PubMed
-
- Miller M, Jaskolski M, Mohana Rao JK, Leis J, Wlodawer A. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature. 1989;337(6207):576–579. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

