Recombinase polymerase amplification (RPA) of CaMV-35S promoter and nos terminator for rapid detection of genetically modified crops
- PMID: 25310647
- PMCID: PMC4227211
- DOI: 10.3390/ijms151018197
Recombinase polymerase amplification (RPA) of CaMV-35S promoter and nos terminator for rapid detection of genetically modified crops
Abstract
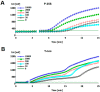
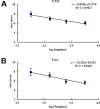
Recombinase polymerase amplification (RPA) is a novel isothermal DNA amplification and detection technology that enables the amplification of DNA within 30 min at a constant temperature of 37-42 °C by simulating in vivo DNA recombination. In this study, based on the regulatory sequence of the cauliflower mosaic virus 35S (CaMV-35S) promoter and the Agrobacterium tumefaciens nopaline synthase gene (nos) terminator, which are widely incorporated in genetically modified (GM) crops, we designed two sets of RPA primers and established a real-time RPA detection method for GM crop screening and detection. This method could reliably detect as few as 100 copies of the target molecule in a sample within 15-25 min. Furthermore, the real-time RPA detection method was successfully used to amplify and detect DNA from samples of four major GM crops (maize, rice, cotton, and soybean). With this novel amplification method, the test time was significantly shortened and the reaction process was simplified; thus, this method represents an effective approach to the rapid detection of GM crops.
Figures
References
-
- James C. Global Status of Commercialized Biotech/GM Crops: 2013. ISAAA; Ithaca, NY, USA: 2013. ISAAA Brief No. 46.
-
- Holst-Jensen A., Rønning S.B., Løvseth A., Berdal K.G. PCR technology for screening and quantification of genetically modified organisms (GMOs) Anal. Bioanal. Chem. 2003;375:985–993. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

