New insights into domestication of carrot from root transcriptome analyses
- PMID: 25311557
- PMCID: PMC4213543
- DOI: 10.1186/1471-2164-15-895
New insights into domestication of carrot from root transcriptome analyses
Abstract
Background: Understanding the molecular basis of domestication can provide insights into the processes of rapid evolution and crop improvement. Here we demonstrated the processes of carrot domestication and identified genes under selection based on transcriptome analyses.
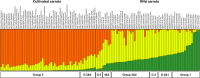
Results: The root transcriptomes of widely differing cultivated and wild carrots were sequenced. A method accounting for sequencing errors was introduced to optimize SNP (single nucleotide polymorphism) discovery. 11,369 SNPs were identified. Of these, 622 (out of 1000 tested SNPs) were validated and used to genotype a large set of cultivated carrot, wild carrot and other wild Daucus carota subspecies, primarily of European origin. Phylogenetic analysis indicated that eastern carrot may originate from Western Asia and western carrot may be selected from eastern carrot. Different wild D. carota subspecies may have contributed to the domestication of cultivated carrot. Genetic diversity was significantly reduced in western cultivars, probably through bottlenecks and selection. However, a high proportion of genetic diversity (more than 85% of the genetic diversity in wild populations) is currently retained in western cultivars. Model simulation indicated high and asymmetric gene flow from wild to cultivated carrots, spontaneously and/or by introgression breeding. Nevertheless, high genetic differentiation exists between cultivated and wild carrots (Fst = 0.295) showing the strong effects of selection. Expression patterns differed radically for some genes between cultivated and wild carrot roots which may be related to changes in root traits. The up-regulation of water-channel-protein gene expression in cultivars might be involved in changing water content and transport in roots. The activated expression of carotenoid-binding-protein genes in cultivars could be related to the high carotenoid accumulation in roots. The silencing of allergen-protein-like genes in cultivated carrot roots suggested strong human selection to reduce allergy. These results suggest that regulatory changes of gene expressions may have played a predominant role in domestication.
Conclusions: Western carrots may originate from eastern carrots. The reduction in genetic diversity in western cultivars due to domestication bottleneck/selection may have been offset by introgression from wild carrot. Differential gene expression patterns between cultivated and wild carrot roots may be a signature of strong selection for favorable cultivation traits.
Figures



Similar articles
-
Genetic structure and domestication of carrot (Daucus carota subsp. sativus) (Apiaceae).Am J Bot. 2013 May;100(5):930-8. doi: 10.3732/ajb.1300055. Epub 2013 Apr 17. Am J Bot. 2013. PMID: 23594914
-
Functional gene polymorphism to reveal species history: the case of the CRTISO gene in cultivated carrots.PLoS One. 2013 Aug 5;8(8):e70801. doi: 10.1371/journal.pone.0070801. Print 2013. PLoS One. 2013. PMID: 23940644 Free PMC article.
-
Carotenoid Presence Is Associated with the Or Gene in Domesticated Carrot.Genetics. 2018 Dec;210(4):1497-1508. doi: 10.1534/genetics.118.301299. Epub 2018 Oct 23. Genetics. 2018. PMID: 30352832 Free PMC article.
-
Pollen-mediated gene flow from wild carrots (Daucus carota L. subsp. carota) affects the production of commercial carrot seeds (Daucus carota L. subsp. sativus) internationally and in New Zealand in the context of climate change: A systematic review.Sci Total Environ. 2024 Jul 10;933:173269. doi: 10.1016/j.scitotenv.2024.173269. Epub 2024 May 14. Sci Total Environ. 2024. PMID: 38754518
-
Biosynthesis of carotenoids in carrot: an underground story comes to light.Arch Biochem Biophys. 2013 Nov 15;539(2):110-6. doi: 10.1016/j.abb.2013.07.009. Epub 2013 Jul 19. Arch Biochem Biophys. 2013. PMID: 23876238 Review.
Cited by
-
Morphological and molecular characterization of variation in common bean (Phaseolus vulgaris L.) germplasm from Azad Jammu and Kashmir, Pakistan.PLoS One. 2022 Apr 26;17(4):e0265817. doi: 10.1371/journal.pone.0265817. eCollection 2022. PLoS One. 2022. PMID: 35472209 Free PMC article.
-
Advances in research on the carrot, an important root vegetable in the Apiaceae family.Hortic Res. 2019 Jun 1;6:69. doi: 10.1038/s41438-019-0150-6. eCollection 2019. Hortic Res. 2019. PMID: 31231527 Free PMC article. Review.
-
Characterization of the watercress (Nasturtium officinale R. Br.; Brassicaceae) transcriptome using RNASeq and identification of candidate genes for important phytonutrient traits linked to human health.BMC Genomics. 2016 May 20;17:378. doi: 10.1186/s12864-016-2704-4. BMC Genomics. 2016. PMID: 27206485 Free PMC article.
-
Transcriptome Analyses Throughout Chili Pepper Fruit Development Reveal Novel Insights into the Domestication Process.Plants (Basel). 2021 Mar 19;10(3):585. doi: 10.3390/plants10030585. Plants (Basel). 2021. PMID: 33808668 Free PMC article.
-
Origin, evolution, breeding, and omics of Apiaceae: a family of vegetables and medicinal plants.Hortic Res. 2022 Apr 11;9:uhac076. doi: 10.1093/hr/uhac076. eCollection 2022. Hortic Res. 2022. PMID: 38239769 Free PMC article.
References
-
- Heywood VH. Relationships and evolution in the Daucus carota complex. Israel J Bot. 1983;32:51–65.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

