Properties of selected mutations and genotypic landscapes under Fisher's geometric model
- PMID: 25311558
- PMCID: PMC4326662
- DOI: 10.1111/evo.12545
Properties of selected mutations and genotypic landscapes under Fisher's geometric model
Abstract
The fitness landscape-the mapping between genotypes and fitness-determines properties of the process of adaptation. Several small genotypic fitness landscapes have recently been built by selecting a handful of beneficial mutations and measuring fitness of all combinations of these mutations. Here, we generate several testable predictions for the properties of these small genotypic landscapes under Fisher's geometric model of adaptation. When the ancestral strain is far from the fitness optimum, we analytically compute the fitness effect of selected mutations and their epistatic interactions. Epistasis may be negative or positive on average depending on the distance of the ancestral genotype to the optimum and whether mutations were independently selected, or coselected in an adaptive walk. Simulations show that genotypic landscapes built from Fisher's model are very close to an additive landscape when the ancestral strain is far from the optimum. However, when it is close to the optimum, a large diversity of landscape with substantial roughness and sign epistasis emerged. Strikingly, small genotypic landscapes built from several replicate adaptive walks on the same underlying landscape were highly variable, suggesting that several realizations of small genotypic landscapes are needed to gain information about the underlying architecture of the fitness landscape.
Keywords: Adaptation; NK model; Rough Mount Fuji; epistasis; experimental evolution; selection.
© 2014 The Author(s). Evolution © 2014 The Society for the Study of Evolution.
Figures

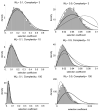
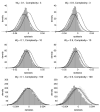




References
References of Appendix
-
- Fisher R. The genetical theory of natural selection. Clarendon Press; 1930.
-
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge: 1983.
-
- Lourenço J, Galtier N, Glémin S. Complexity, pleiotropy, and the fitness effect of mutations. Evolution. 2011;65(6):1559–1571. - PubMed
-
- Martin G, Elena SF, Lenormand T. Distributions of epistasis in microbes fit predictions from a fitness landscape model. Nature genetics. 2007;39(4):555–560. - PubMed
-
- Martin G, Lenormand T. A general multivariate extension of Fisher’s geometrical model and the distribution of mutation fitness effects across species. Evolution. 2006;60(5):893–907. - PubMed
References
-
- Achaz G, Rodriguez-Verdugo A, Gaut BS, Tenaillon O. Ecological Genomics. Springer; 2014. The reproducibility of adaptation in the light of experimental evolution with whole genome sequencing; pp. 211–231. - PubMed
-
- Chevin L-M, Decorzent G, Lenormand T. Niche dimensionality and the genetics of ecological speciation. Evolution. 2014;68(5):1244–1256. - PubMed
-
- Chevin L, Martin G, Lenormand T. Fisher’s model and the genomics of adaptation: restricted pleiotropy, heterogeneous mutation, and parallel evolution. Evolution. 2010;64(11):3213–3231. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
