Machine learning framework for early MRI-based Alzheimer's conversion prediction in MCI subjects
- PMID: 25312773
- PMCID: PMC5957071
- DOI: 10.1016/j.neuroimage.2014.10.002
Machine learning framework for early MRI-based Alzheimer's conversion prediction in MCI subjects
Abstract
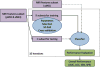
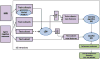
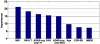
Mild cognitive impairment (MCI) is a transitional stage between age-related cognitive decline and Alzheimer's disease (AD). For the effective treatment of AD, it would be important to identify MCI patients at high risk for conversion to AD. In this study, we present a novel magnetic resonance imaging (MRI)-based method for predicting the MCI-to-AD conversion from one to three years before the clinical diagnosis. First, we developed a novel MRI biomarker of MCI-to-AD conversion using semi-supervised learning and then integrated it with age and cognitive measures about the subjects using a supervised learning algorithm resulting in what we call the aggregate biomarker. The novel characteristics of the methods for learning the biomarkers are as follows: 1) We used a semi-supervised learning method (low density separation) for the construction of MRI biomarker as opposed to more typical supervised methods; 2) We performed a feature selection on MRI data from AD subjects and normal controls without using data from MCI subjects via regularized logistic regression; 3) We removed the aging effects from the MRI data before the classifier training to prevent possible confounding between AD and age related atrophies; and 4) We constructed the aggregate biomarker by first learning a separate MRI biomarker and then combining it with age and cognitive measures about the MCI subjects at the baseline by applying a random forest classifier. We experimentally demonstrated the added value of these novel characteristics in predicting the MCI-to-AD conversion on data obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database. With the ADNI data, the MRI biomarker achieved a 10-fold cross-validated area under the receiver operating characteristic curve (AUC) of 0.7661 in discriminating progressive MCI patients (pMCI) from stable MCI patients (sMCI). Our aggregate biomarker based on MRI data together with baseline cognitive measurements and age achieved a 10-fold cross-validated AUC score of 0.9020 in discriminating pMCI from sMCI. The results presented in this study demonstrate the potential of the suggested approach for early AD diagnosis and an important role of MRI in the MCI-to-AD conversion prediction. However, it is evident based on our results that combining MRI data with cognitive test results improved the accuracy of the MCI-to-AD conversion prediction.
Keywords: ADNI; Alzheimer's disease; Classification; Early diagnosis; Feature selection; Low density separation; Magnetic resonance imaging; Mild cognitive impairment; Semi-supervised learning; Support vector machine.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

