Sequence selectivity of macrolide-induced translational attenuation
- PMID: 25313041
- PMCID: PMC4217412
- DOI: 10.1073/pnas.1410356111
Sequence selectivity of macrolide-induced translational attenuation
Abstract
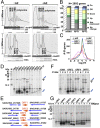
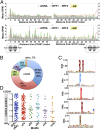
The prevailing "plug-in-the-bottle" model suggests that macrolide antibiotics inhibit translation by binding inside the ribosome tunnel and indiscriminately arresting the elongation of every nascent polypeptide after the synthesis of six to eight amino acids. To test this model, we performed a genome-wide analysis of translation in azithromycin-treated Staphylococcus aureus. In contrast to earlier predictions, we found that the macrolide does not preferentially induce ribosome stalling near the 5' end of mRNAs, but rather acts at specific stalling sites that are scattered throughout the entire coding region. These sites are highly enriched in prolines and charged residues and are strikingly similar to other ligand-independent ribosome stalling motifs. Interestingly, the addition of structurally related macrolides had dramatically different effects on stalling efficiency. Our data suggest that ribosome stalling can occur at a surprisingly large number of low-complexity motifs in a fashion that depends only on a few arrest-inducing residues and the presence of a small molecule inducer.
Keywords: Staphylococcus aureus; antibiotic; ribosome stalling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289(5481):920–930. - PubMed
-
- Bhushan S, et al. alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol. 2010;17(3):313–317. - PubMed
-
- Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44(23):8230–8243. - PubMed
-
- Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol Cell. 2006;22(5):587–598. - PubMed
-
- Ito K, Chiba S. Arrest peptides: Cis-acting modulators of translation. Annu Rev Biochem. 2013;82:171–202. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

