RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer
- PMID: 25313140
- PMCID: PMC4259446
- DOI: 10.18632/oncotarget.2476
RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer
Abstract
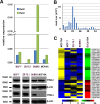
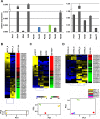
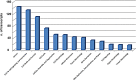
PIWI-interacting small non-coding RNAs (piRNAs) are genetic and epigenetic regulatory factors in germline cells, where they maintain genome stability, are involved in RNA silencing and regulate gene expression. We found that the piRNA biogenesis and effector pathway are present in human breast cancer (BC) cells and, analyzing smallRNA-Seq data generated from BC cell lines and tumor biopsies, we identified >100 BC piRNAs, including some very abundant and/or differentially expressed in mammary epithelial compared to BC cells, where this was influenced by estrogen or estrogen receptor β, and in cancer respect to normal breast tissues. A search for mRNAs targeted by the BC piRNome revealed that eight piRNAs showing a specific expression pattern in breast tumors target key cancer cell pathways. Evidence of an active piRNA pathway in BC suggests that these small non-coding RNAs do exert transcriptional and post-transcriptional gene regulatory actions also in cancer cells.
Conflict of interest statement
The Authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

